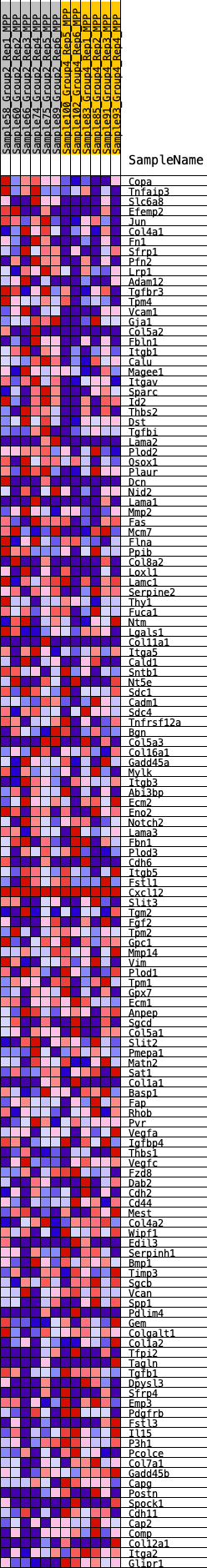
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
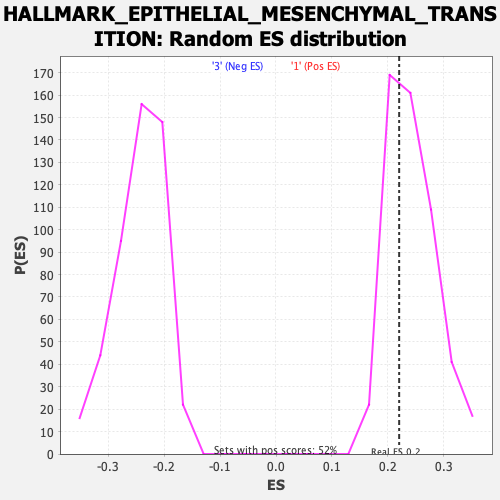
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.22051422 |
| Normalized Enrichment Score (NES) | 0.90971965 |
| Nominal p-value | 0.65317917 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Copa | 602 | 0.572 | -0.0133 | Yes |
| 2 | Tnfaip3 | 781 | 0.533 | -0.0059 | Yes |
| 3 | Slc6a8 | 835 | 0.525 | 0.0077 | Yes |
| 4 | Efemp2 | 907 | 0.510 | 0.0200 | Yes |
| 5 | Jun | 963 | 0.503 | 0.0328 | Yes |
| 6 | Col4a1 | 1060 | 0.487 | 0.0430 | Yes |
| 7 | Fn1 | 1090 | 0.484 | 0.0565 | Yes |
| 8 | Sfrp1 | 1138 | 0.476 | 0.0689 | Yes |
| 9 | Pfn2 | 1174 | 0.470 | 0.0817 | Yes |
| 10 | Lrp1 | 1468 | 0.433 | 0.0801 | Yes |
| 11 | Adam12 | 1475 | 0.431 | 0.0932 | Yes |
| 12 | Tgfbr3 | 1610 | 0.416 | 0.0992 | Yes |
| 13 | Tpm4 | 1627 | 0.414 | 0.1113 | Yes |
| 14 | Vcam1 | 1741 | 0.403 | 0.1180 | Yes |
| 15 | Gja1 | 1782 | 0.399 | 0.1283 | Yes |
| 16 | Col5a2 | 1813 | 0.396 | 0.1391 | Yes |
| 17 | Fbln1 | 1826 | 0.395 | 0.1508 | Yes |
| 18 | Itgb1 | 2048 | 0.372 | 0.1509 | Yes |
| 19 | Calu | 2115 | 0.363 | 0.1588 | Yes |
| 20 | Magee1 | 2200 | 0.357 | 0.1656 | Yes |
| 21 | Itgav | 2434 | 0.338 | 0.1641 | Yes |
| 22 | Sparc | 2479 | 0.335 | 0.1722 | Yes |
| 23 | Id2 | 2550 | 0.330 | 0.1789 | Yes |
| 24 | Thbs2 | 2597 | 0.326 | 0.1867 | Yes |
| 25 | Dst | 2658 | 0.322 | 0.1936 | Yes |
| 26 | Tgfbi | 2804 | 0.311 | 0.1958 | Yes |
| 27 | Lama2 | 2957 | 0.301 | 0.1973 | Yes |
| 28 | Plod2 | 2978 | 0.301 | 0.2056 | Yes |
| 29 | Qsox1 | 2999 | 0.300 | 0.2139 | Yes |
| 30 | Plaur | 3051 | 0.297 | 0.2205 | Yes |
| 31 | Dcn | 3341 | 0.278 | 0.2142 | No |
| 32 | Nid2 | 4028 | 0.238 | 0.1862 | No |
| 33 | Lama1 | 4162 | 0.229 | 0.1865 | No |
| 34 | Mmp2 | 4287 | 0.222 | 0.1870 | No |
| 35 | Fas | 4319 | 0.220 | 0.1923 | No |
| 36 | Mcm7 | 4783 | 0.198 | 0.1745 | No |
| 37 | Flna | 4907 | 0.192 | 0.1742 | No |
| 38 | Ppib | 5480 | 0.163 | 0.1497 | No |
| 39 | Col8a2 | 5528 | 0.161 | 0.1523 | No |
| 40 | Loxl1 | 5656 | 0.155 | 0.1506 | No |
| 41 | Lamc1 | 5809 | 0.148 | 0.1473 | No |
| 42 | Serpine2 | 5984 | 0.139 | 0.1427 | No |
| 43 | Thy1 | 6023 | 0.137 | 0.1450 | No |
| 44 | Fuca1 | 6086 | 0.134 | 0.1460 | No |
| 45 | Ntm | 6244 | 0.127 | 0.1418 | No |
| 46 | Lgals1 | 6296 | 0.125 | 0.1431 | No |
| 47 | Col11a1 | 6365 | 0.122 | 0.1434 | No |
| 48 | Itga5 | 6416 | 0.122 | 0.1446 | No |
| 49 | Cald1 | 6443 | 0.120 | 0.1470 | No |
| 50 | Sntb1 | 6698 | 0.109 | 0.1372 | No |
| 51 | Nt5e | 6887 | 0.099 | 0.1306 | No |
| 52 | Sdc1 | 7084 | 0.090 | 0.1233 | No |
| 53 | Cadm1 | 7294 | 0.082 | 0.1151 | No |
| 54 | Sdc4 | 7430 | 0.077 | 0.1105 | No |
| 55 | Tnfrsf12a | 7527 | 0.074 | 0.1079 | No |
| 56 | Bgn | 7659 | 0.069 | 0.1033 | No |
| 57 | Col5a3 | 7676 | 0.068 | 0.1046 | No |
| 58 | Col16a1 | 7713 | 0.067 | 0.1048 | No |
| 59 | Gadd45a | 7891 | 0.060 | 0.0975 | No |
| 60 | Mylk | 7920 | 0.058 | 0.0979 | No |
| 61 | Itgb3 | 8047 | 0.053 | 0.0930 | No |
| 62 | Abi3bp | 8075 | 0.052 | 0.0932 | No |
| 63 | Ecm2 | 8132 | 0.050 | 0.0919 | No |
| 64 | Eno2 | 8150 | 0.049 | 0.0925 | No |
| 65 | Notch2 | 8243 | 0.045 | 0.0892 | No |
| 66 | Lama3 | 8297 | 0.043 | 0.0878 | No |
| 67 | Fbn1 | 8607 | 0.030 | 0.0728 | No |
| 68 | Plod3 | 8771 | 0.024 | 0.0651 | No |
| 69 | Cdh6 | 8905 | 0.018 | 0.0588 | No |
| 70 | Itgb5 | 9228 | 0.007 | 0.0424 | No |
| 71 | Fstl1 | 9236 | 0.007 | 0.0423 | No |
| 72 | Cxcl12 | 9445 | 0.000 | 0.0315 | No |
| 73 | Slit3 | 9720 | -0.009 | 0.0177 | No |
| 74 | Tgm2 | 9755 | -0.010 | 0.0162 | No |
| 75 | Fgf2 | 10082 | -0.023 | 0.0001 | No |
| 76 | Tpm2 | 10128 | -0.025 | -0.0014 | No |
| 77 | Gpc1 | 10397 | -0.037 | -0.0141 | No |
| 78 | Mmp14 | 10403 | -0.037 | -0.0132 | No |
| 79 | Vim | 10510 | -0.041 | -0.0174 | No |
| 80 | Plod1 | 10737 | -0.049 | -0.0275 | No |
| 81 | Tpm1 | 10849 | -0.054 | -0.0316 | No |
| 82 | Gpx7 | 10935 | -0.057 | -0.0342 | No |
| 83 | Ecm1 | 10989 | -0.059 | -0.0351 | No |
| 84 | Anpep | 11008 | -0.060 | -0.0341 | No |
| 85 | Sgcd | 11057 | -0.062 | -0.0347 | No |
| 86 | Col5a1 | 11140 | -0.066 | -0.0369 | No |
| 87 | Slit2 | 11443 | -0.077 | -0.0501 | No |
| 88 | Pmepa1 | 11737 | -0.090 | -0.0624 | No |
| 89 | Matn2 | 11859 | -0.096 | -0.0656 | No |
| 90 | Sat1 | 12161 | -0.109 | -0.0778 | No |
| 91 | Col1a1 | 12334 | -0.116 | -0.0831 | No |
| 92 | Basp1 | 12500 | -0.122 | -0.0878 | No |
| 93 | Fap | 12668 | -0.127 | -0.0925 | No |
| 94 | Rhob | 12685 | -0.128 | -0.0893 | No |
| 95 | Pvr | 12822 | -0.133 | -0.0922 | No |
| 96 | Vegfa | 13223 | -0.151 | -0.1081 | No |
| 97 | Igfbp4 | 13439 | -0.162 | -0.1142 | No |
| 98 | Thbs1 | 13449 | -0.163 | -0.1096 | No |
| 99 | Vegfc | 13552 | -0.167 | -0.1097 | No |
| 100 | Fzd8 | 13878 | -0.181 | -0.1208 | No |
| 101 | Dab2 | 13934 | -0.184 | -0.1179 | No |
| 102 | Cdh2 | 14183 | -0.196 | -0.1247 | No |
| 103 | Cd44 | 14250 | -0.199 | -0.1219 | No |
| 104 | Mest | 14363 | -0.204 | -0.1213 | No |
| 105 | Col4a2 | 14579 | -0.215 | -0.1257 | No |
| 106 | Wipf1 | 14757 | -0.223 | -0.1279 | No |
| 107 | Edil3 | 14895 | -0.231 | -0.1278 | No |
| 108 | Serpinh1 | 14907 | -0.232 | -0.1212 | No |
| 109 | Bmp1 | 14909 | -0.232 | -0.1140 | No |
| 110 | Timp3 | 14954 | -0.234 | -0.1090 | No |
| 111 | Sgcb | 15011 | -0.237 | -0.1045 | No |
| 112 | Vcan | 15051 | -0.240 | -0.0991 | No |
| 113 | Spp1 | 15092 | -0.242 | -0.0936 | No |
| 114 | Pdlim4 | 15108 | -0.242 | -0.0869 | No |
| 115 | Gem | 15191 | -0.246 | -0.0834 | No |
| 116 | Colgalt1 | 15445 | -0.260 | -0.0884 | No |
| 117 | Col1a2 | 15646 | -0.271 | -0.0903 | No |
| 118 | Tfpi2 | 16084 | -0.297 | -0.1036 | No |
| 119 | Tagln | 16180 | -0.301 | -0.0991 | No |
| 120 | Tgfb1 | 16282 | -0.307 | -0.0948 | No |
| 121 | Dpysl3 | 16336 | -0.311 | -0.0878 | No |
| 122 | Sfrp4 | 16342 | -0.311 | -0.0784 | No |
| 123 | Emp3 | 16377 | -0.314 | -0.0704 | No |
| 124 | Pdgfrb | 16728 | -0.335 | -0.0780 | No |
| 125 | Fstl3 | 16899 | -0.348 | -0.0760 | No |
| 126 | Il15 | 16988 | -0.354 | -0.0695 | No |
| 127 | P3h1 | 17184 | -0.368 | -0.0681 | No |
| 128 | Pcolce | 17463 | -0.390 | -0.0703 | No |
| 129 | Col7a1 | 17816 | -0.423 | -0.0754 | No |
| 130 | Gadd45b | 17861 | -0.428 | -0.0643 | No |
| 131 | Capg | 17995 | -0.442 | -0.0574 | No |
| 132 | Postn | 18109 | -0.454 | -0.0491 | No |
| 133 | Spock1 | 18135 | -0.456 | -0.0362 | No |
| 134 | Cdh11 | 18207 | -0.465 | -0.0254 | No |
| 135 | Cap2 | 18453 | -0.501 | -0.0225 | No |
| 136 | Comp | 18501 | -0.508 | -0.0091 | No |
| 137 | Col12a1 | 18629 | -0.528 | 0.0008 | No |
| 138 | Itga2 | 18846 | -0.570 | 0.0073 | No |
| 139 | Glipr1 | 19413 | -0.867 | 0.0051 | No |