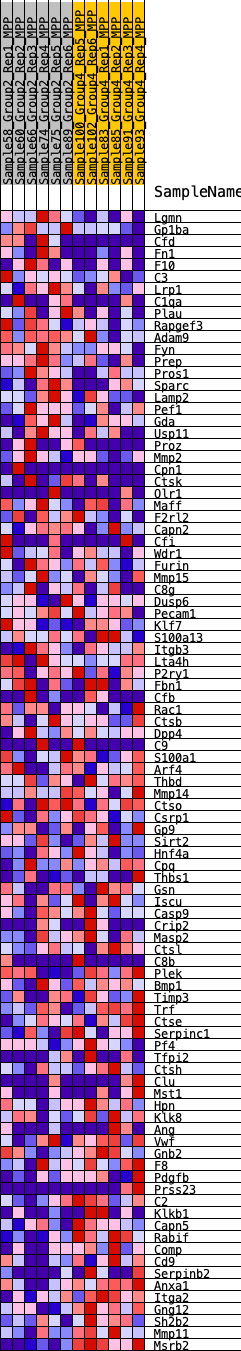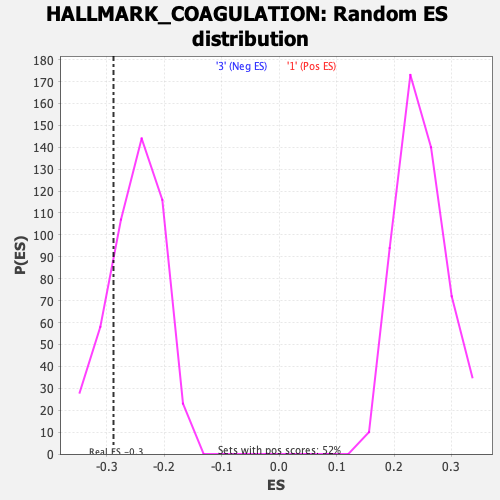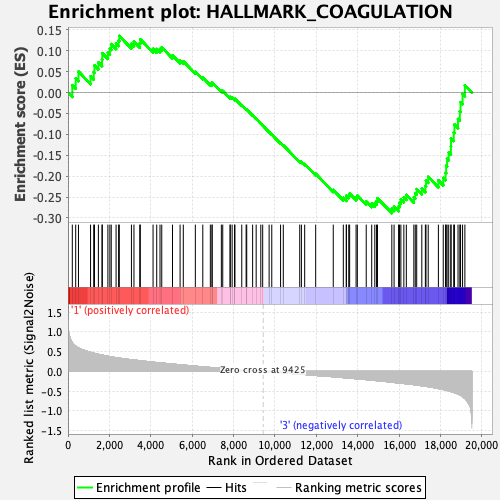
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.28845048 |
| Normalized Enrichment Score (NES) | -1.1583945 |
| Nominal p-value | 0.1869748 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.959 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lgmn | 208 | 0.726 | 0.0175 | No |
| 2 | Gp1ba | 372 | 0.639 | 0.0339 | No |
| 3 | Cfd | 506 | 0.593 | 0.0500 | No |
| 4 | Fn1 | 1090 | 0.484 | 0.0388 | No |
| 5 | F10 | 1243 | 0.461 | 0.0488 | No |
| 6 | C3 | 1279 | 0.457 | 0.0648 | No |
| 7 | Lrp1 | 1468 | 0.433 | 0.0719 | No |
| 8 | C1qa | 1643 | 0.412 | 0.0789 | No |
| 9 | Plau | 1658 | 0.411 | 0.0941 | No |
| 10 | Rapgef3 | 1928 | 0.384 | 0.0952 | No |
| 11 | Adam9 | 2023 | 0.374 | 0.1049 | No |
| 12 | Fyn | 2089 | 0.367 | 0.1158 | No |
| 13 | Prep | 2323 | 0.346 | 0.1172 | No |
| 14 | Pros1 | 2445 | 0.337 | 0.1241 | No |
| 15 | Sparc | 2479 | 0.335 | 0.1354 | No |
| 16 | Lamp2 | 3065 | 0.296 | 0.1168 | No |
| 17 | Pef1 | 3187 | 0.288 | 0.1217 | No |
| 18 | Gda | 3468 | 0.273 | 0.1179 | No |
| 19 | Usp11 | 3494 | 0.271 | 0.1271 | No |
| 20 | Proz | 4110 | 0.233 | 0.1044 | No |
| 21 | Mmp2 | 4287 | 0.222 | 0.1040 | No |
| 22 | Cpn1 | 4446 | 0.216 | 0.1042 | No |
| 23 | Ctsk | 4530 | 0.212 | 0.1082 | No |
| 24 | Olr1 | 5049 | 0.186 | 0.0888 | No |
| 25 | Maff | 5416 | 0.167 | 0.0764 | No |
| 26 | F2rl2 | 5571 | 0.159 | 0.0746 | No |
| 27 | Capn2 | 6153 | 0.132 | 0.0498 | No |
| 28 | Cfi | 6513 | 0.117 | 0.0359 | No |
| 29 | Wdr1 | 6878 | 0.099 | 0.0210 | No |
| 30 | Furin | 6959 | 0.096 | 0.0206 | No |
| 31 | Mmp15 | 6965 | 0.095 | 0.0241 | No |
| 32 | C8g | 7410 | 0.078 | 0.0042 | No |
| 33 | Dusp6 | 7477 | 0.076 | 0.0038 | No |
| 34 | Pecam1 | 7831 | 0.063 | -0.0120 | No |
| 35 | Klf7 | 7839 | 0.062 | -0.0099 | No |
| 36 | S100a13 | 7931 | 0.058 | -0.0124 | No |
| 37 | Itgb3 | 8047 | 0.053 | -0.0162 | No |
| 38 | Lta4h | 8066 | 0.052 | -0.0151 | No |
| 39 | P2ry1 | 8396 | 0.039 | -0.0305 | No |
| 40 | Fbn1 | 8607 | 0.030 | -0.0402 | No |
| 41 | Cfb | 8636 | 0.029 | -0.0405 | No |
| 42 | Rac1 | 8921 | 0.017 | -0.0545 | No |
| 43 | Ctsb | 9092 | 0.012 | -0.0627 | No |
| 44 | Dpp4 | 9316 | 0.004 | -0.0741 | No |
| 45 | C9 | 9407 | 0.000 | -0.0787 | No |
| 46 | S100a1 | 9718 | -0.009 | -0.0943 | No |
| 47 | Arf4 | 9846 | -0.014 | -0.1003 | No |
| 48 | Thbd | 10271 | -0.031 | -0.1209 | No |
| 49 | Mmp14 | 10403 | -0.037 | -0.1262 | No |
| 50 | Ctso | 11197 | -0.068 | -0.1644 | No |
| 51 | Csrp1 | 11278 | -0.070 | -0.1658 | No |
| 52 | Gp9 | 11436 | -0.077 | -0.1709 | No |
| 53 | Sirt2 | 11968 | -0.100 | -0.1944 | No |
| 54 | Hnf4a | 12817 | -0.133 | -0.2329 | No |
| 55 | Cpq | 13303 | -0.155 | -0.2518 | No |
| 56 | Thbs1 | 13449 | -0.163 | -0.2530 | No |
| 57 | Gsn | 13450 | -0.163 | -0.2467 | No |
| 58 | Iscu | 13566 | -0.167 | -0.2461 | No |
| 59 | Casp9 | 13609 | -0.170 | -0.2417 | No |
| 60 | Crip2 | 13920 | -0.183 | -0.2505 | No |
| 61 | Masp2 | 13981 | -0.186 | -0.2464 | No |
| 62 | Ctsl | 14410 | -0.206 | -0.2604 | No |
| 63 | C8b | 14675 | -0.220 | -0.2655 | No |
| 64 | Plek | 14825 | -0.227 | -0.2644 | No |
| 65 | Bmp1 | 14909 | -0.232 | -0.2596 | No |
| 66 | Timp3 | 14954 | -0.234 | -0.2528 | No |
| 67 | Trf | 15647 | -0.271 | -0.2779 | Yes |
| 68 | Ctse | 15756 | -0.278 | -0.2727 | Yes |
| 69 | Serpinc1 | 15973 | -0.292 | -0.2725 | Yes |
| 70 | Pf4 | 16019 | -0.294 | -0.2634 | Yes |
| 71 | Tfpi2 | 16084 | -0.297 | -0.2551 | Yes |
| 72 | Ctsh | 16228 | -0.304 | -0.2507 | Yes |
| 73 | Clu | 16348 | -0.311 | -0.2448 | Yes |
| 74 | Mst1 | 16711 | -0.334 | -0.2504 | Yes |
| 75 | Hpn | 16780 | -0.338 | -0.2408 | Yes |
| 76 | Klk8 | 16849 | -0.344 | -0.2310 | Yes |
| 77 | Ang | 17096 | -0.361 | -0.2296 | Yes |
| 78 | Vwf | 17268 | -0.374 | -0.2239 | Yes |
| 79 | Gnb2 | 17297 | -0.376 | -0.2108 | Yes |
| 80 | F8 | 17405 | -0.385 | -0.2013 | Yes |
| 81 | Pdgfb | 17897 | -0.432 | -0.2099 | Yes |
| 82 | Prss23 | 18132 | -0.456 | -0.2042 | Yes |
| 83 | C2 | 18243 | -0.470 | -0.1916 | Yes |
| 84 | Klkb1 | 18273 | -0.473 | -0.1748 | Yes |
| 85 | Capn5 | 18317 | -0.479 | -0.1584 | Yes |
| 86 | Rabif | 18395 | -0.491 | -0.1433 | Yes |
| 87 | Comp | 18501 | -0.508 | -0.1290 | Yes |
| 88 | Cd9 | 18507 | -0.509 | -0.1095 | Yes |
| 89 | Serpinb2 | 18638 | -0.530 | -0.0956 | Yes |
| 90 | Anxa1 | 18673 | -0.537 | -0.0765 | Yes |
| 91 | Itga2 | 18846 | -0.570 | -0.0633 | Yes |
| 92 | Gng12 | 18933 | -0.590 | -0.0448 | Yes |
| 93 | Sh2b2 | 18968 | -0.601 | -0.0232 | Yes |
| 94 | Mmp11 | 19065 | -0.635 | -0.0035 | Yes |
| 95 | Msrb2 | 19177 | -0.682 | 0.0173 | Yes |