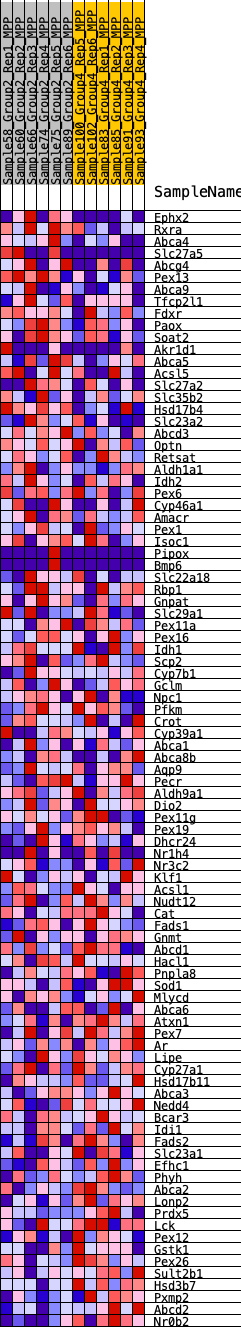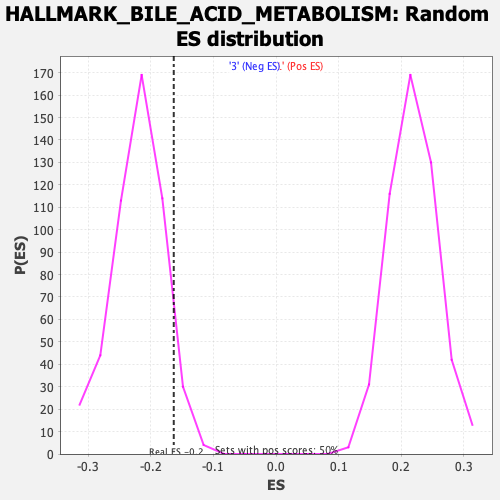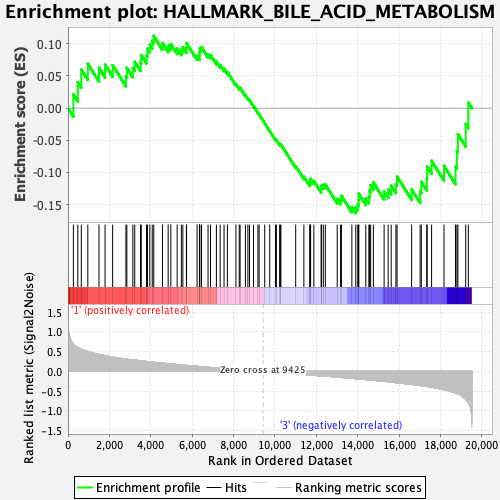
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.16318712 |
| Normalized Enrichment Score (NES) | -0.74528354 |
| Nominal p-value | 0.9354839 |
| FDR q-value | 0.89236903 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ephx2 | 260 | 0.691 | 0.0212 | No |
| 2 | Rxra | 475 | 0.603 | 0.0404 | No |
| 3 | Abca4 | 643 | 0.561 | 0.0599 | No |
| 4 | Slc27a5 | 958 | 0.503 | 0.0689 | No |
| 5 | Abcg4 | 1498 | 0.429 | 0.0626 | No |
| 6 | Pex13 | 1791 | 0.398 | 0.0675 | No |
| 7 | Abca9 | 2157 | 0.360 | 0.0667 | No |
| 8 | Tfcp2l1 | 2800 | 0.311 | 0.0492 | No |
| 9 | Fdxr | 2843 | 0.308 | 0.0625 | No |
| 10 | Paox | 3133 | 0.291 | 0.0622 | No |
| 11 | Soat2 | 3224 | 0.285 | 0.0718 | No |
| 12 | Akr1d1 | 3502 | 0.270 | 0.0711 | No |
| 13 | Abca5 | 3540 | 0.267 | 0.0825 | No |
| 14 | Acsl5 | 3795 | 0.251 | 0.0820 | No |
| 15 | Slc27a2 | 3841 | 0.249 | 0.0922 | No |
| 16 | Slc35b2 | 3953 | 0.241 | 0.0985 | No |
| 17 | Hsd17b4 | 4070 | 0.235 | 0.1043 | No |
| 18 | Slc23a2 | 4143 | 0.231 | 0.1121 | No |
| 19 | Abcd3 | 4564 | 0.210 | 0.1010 | No |
| 20 | Optn | 4839 | 0.195 | 0.0967 | No |
| 21 | Retsat | 4969 | 0.189 | 0.0995 | No |
| 22 | Aldh1a1 | 5274 | 0.174 | 0.0925 | No |
| 23 | Idh2 | 5476 | 0.164 | 0.0904 | No |
| 24 | Pex6 | 5550 | 0.161 | 0.0947 | No |
| 25 | Cyp46a1 | 5722 | 0.153 | 0.0935 | No |
| 26 | Amacr | 5729 | 0.152 | 0.1008 | No |
| 27 | Pex1 | 6237 | 0.128 | 0.0811 | No |
| 28 | Isoc1 | 6352 | 0.122 | 0.0813 | No |
| 29 | Pipox | 6359 | 0.122 | 0.0871 | No |
| 30 | Bmp6 | 6369 | 0.122 | 0.0928 | No |
| 31 | Slc22a18 | 6452 | 0.120 | 0.0945 | No |
| 32 | Rbp1 | 6767 | 0.105 | 0.0836 | No |
| 33 | Gnpat | 6885 | 0.099 | 0.0825 | No |
| 34 | Slc29a1 | 7171 | 0.087 | 0.0722 | No |
| 35 | Pex11a | 7351 | 0.080 | 0.0670 | No |
| 36 | Pex16 | 7540 | 0.073 | 0.0610 | No |
| 37 | Idh1 | 7702 | 0.067 | 0.0560 | No |
| 38 | Scp2 | 8115 | 0.051 | 0.0374 | No |
| 39 | Cyp7b1 | 8275 | 0.044 | 0.0314 | No |
| 40 | Gclm | 8315 | 0.043 | 0.0315 | No |
| 41 | Npc1 | 8575 | 0.031 | 0.0197 | No |
| 42 | Pfkm | 8692 | 0.027 | 0.0151 | No |
| 43 | Crot | 8770 | 0.024 | 0.0123 | No |
| 44 | Cyp39a1 | 8965 | 0.016 | 0.0031 | No |
| 45 | Abca1 | 9178 | 0.009 | -0.0073 | No |
| 46 | Abca8b | 9233 | 0.007 | -0.0098 | No |
| 47 | Aqp9 | 9505 | -0.001 | -0.0237 | No |
| 48 | Pecr | 9747 | -0.010 | -0.0356 | No |
| 49 | Aldh9a1 | 10024 | -0.022 | -0.0487 | No |
| 50 | Dio2 | 10069 | -0.023 | -0.0498 | No |
| 51 | Pex11g | 10227 | -0.030 | -0.0564 | No |
| 52 | Pex19 | 10258 | -0.031 | -0.0564 | No |
| 53 | Dhcr24 | 10287 | -0.032 | -0.0563 | No |
| 54 | Nr1h4 | 11002 | -0.060 | -0.0900 | No |
| 55 | Nr3c2 | 11397 | -0.075 | -0.1066 | No |
| 56 | Klf1 | 11679 | -0.087 | -0.1167 | No |
| 57 | Acsl1 | 11707 | -0.089 | -0.1136 | No |
| 58 | Nudt12 | 11723 | -0.089 | -0.1099 | No |
| 59 | Cat | 11880 | -0.097 | -0.1131 | No |
| 60 | Fads1 | 12235 | -0.111 | -0.1258 | No |
| 61 | Gnmt | 12242 | -0.112 | -0.1205 | No |
| 62 | Abcd1 | 12336 | -0.116 | -0.1195 | No |
| 63 | Hacl1 | 12432 | -0.119 | -0.1184 | No |
| 64 | Pnpla8 | 13009 | -0.142 | -0.1410 | No |
| 65 | Sod1 | 13171 | -0.149 | -0.1418 | No |
| 66 | Mlycd | 13211 | -0.151 | -0.1363 | No |
| 67 | Abca6 | 13716 | -0.174 | -0.1536 | No |
| 68 | Atxn1 | 13904 | -0.183 | -0.1541 | Yes |
| 69 | Pex7 | 13999 | -0.187 | -0.1495 | Yes |
| 70 | Ar | 14043 | -0.189 | -0.1423 | Yes |
| 71 | Lipe | 14049 | -0.190 | -0.1330 | Yes |
| 72 | Cyp27a1 | 14388 | -0.205 | -0.1402 | Yes |
| 73 | Hsd17b11 | 14538 | -0.213 | -0.1372 | Yes |
| 74 | Abca3 | 14572 | -0.215 | -0.1281 | Yes |
| 75 | Nedd4 | 14623 | -0.217 | -0.1198 | Yes |
| 76 | Bcar3 | 14755 | -0.222 | -0.1154 | Yes |
| 77 | Idi1 | 15274 | -0.251 | -0.1296 | Yes |
| 78 | Fads2 | 15471 | -0.261 | -0.1266 | Yes |
| 79 | Slc23a1 | 15616 | -0.270 | -0.1205 | Yes |
| 80 | Efhc1 | 15845 | -0.284 | -0.1180 | Yes |
| 81 | Phyh | 15900 | -0.287 | -0.1064 | Yes |
| 82 | Abca2 | 16604 | -0.327 | -0.1263 | Yes |
| 83 | Lonp2 | 17015 | -0.356 | -0.1296 | Yes |
| 84 | Prdx5 | 17072 | -0.360 | -0.1145 | Yes |
| 85 | Lck | 17337 | -0.379 | -0.1091 | Yes |
| 86 | Pex12 | 17352 | -0.380 | -0.0908 | Yes |
| 87 | Gstk1 | 17569 | -0.400 | -0.0819 | Yes |
| 88 | Pex26 | 18168 | -0.459 | -0.0897 | Yes |
| 89 | Sult2b1 | 18724 | -0.545 | -0.0910 | Yes |
| 90 | Hsd3b7 | 18791 | -0.559 | -0.0664 | Yes |
| 91 | Pxmp2 | 18840 | -0.568 | -0.0405 | Yes |
| 92 | Abcd2 | 19217 | -0.701 | -0.0248 | Yes |
| 93 | Nr0b2 | 19344 | -0.797 | 0.0087 | Yes |