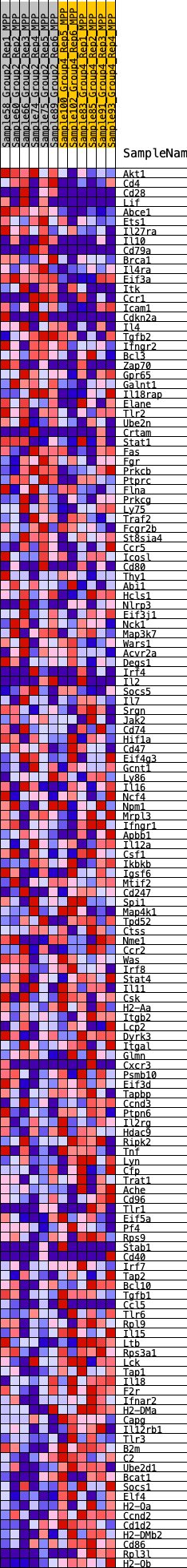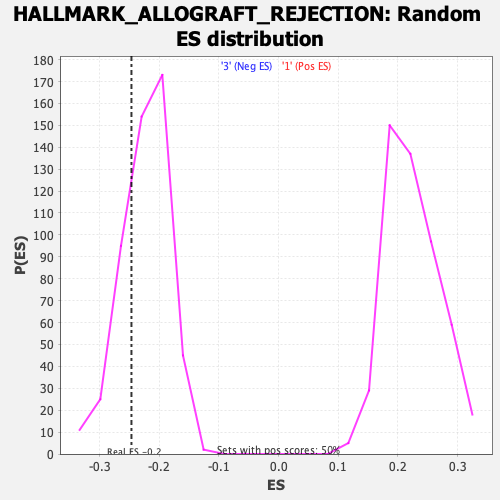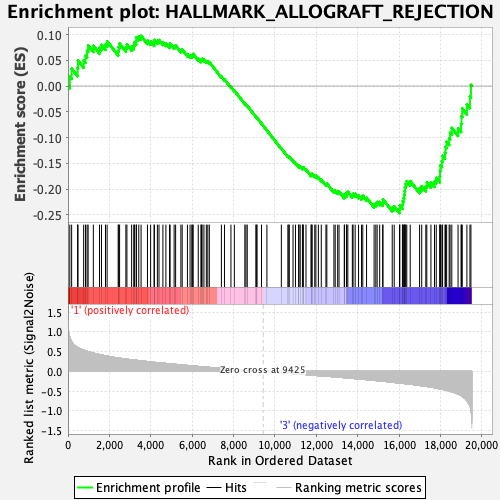
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group4.MPP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.24677393 |
| Normalized Enrichment Score (NES) | -1.1063738 |
| Nominal p-value | 0.26138613 |
| FDR q-value | 0.9855959 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 88 | 0.870 | 0.0183 | No |
| 2 | Cd4 | 176 | 0.756 | 0.0337 | No |
| 3 | Cd28 | 463 | 0.607 | 0.0349 | No |
| 4 | Lif | 483 | 0.601 | 0.0497 | No |
| 5 | Abce1 | 756 | 0.537 | 0.0497 | No |
| 6 | Ets1 | 844 | 0.524 | 0.0590 | No |
| 7 | Il27ra | 921 | 0.508 | 0.0684 | No |
| 8 | Il10 | 972 | 0.502 | 0.0790 | No |
| 9 | Cd79a | 1224 | 0.463 | 0.0782 | No |
| 10 | Brca1 | 1523 | 0.427 | 0.0741 | No |
| 11 | Il4ra | 1618 | 0.415 | 0.0801 | No |
| 12 | Eif3a | 1811 | 0.396 | 0.0806 | No |
| 13 | Itk | 1890 | 0.389 | 0.0868 | No |
| 14 | Ccr1 | 2424 | 0.339 | 0.0682 | No |
| 15 | Icam1 | 2452 | 0.337 | 0.0756 | No |
| 16 | Cdkn2a | 2493 | 0.334 | 0.0824 | No |
| 17 | Il4 | 2797 | 0.311 | 0.0749 | No |
| 18 | Tgfb2 | 2846 | 0.308 | 0.0805 | No |
| 19 | Ifngr2 | 3075 | 0.296 | 0.0765 | No |
| 20 | Bcl3 | 3179 | 0.288 | 0.0788 | No |
| 21 | Zap70 | 3209 | 0.286 | 0.0848 | No |
| 22 | Gpr65 | 3296 | 0.281 | 0.0877 | No |
| 23 | Galnt1 | 3304 | 0.281 | 0.0947 | No |
| 24 | Il18rap | 3420 | 0.275 | 0.0960 | No |
| 25 | Elane | 3531 | 0.268 | 0.0974 | No |
| 26 | Tlr2 | 3839 | 0.249 | 0.0881 | No |
| 27 | Ube2n | 3992 | 0.239 | 0.0865 | No |
| 28 | Crtam | 4161 | 0.229 | 0.0839 | No |
| 29 | Stat1 | 4171 | 0.229 | 0.0894 | No |
| 30 | Fas | 4319 | 0.220 | 0.0876 | No |
| 31 | Fgr | 4397 | 0.218 | 0.0894 | No |
| 32 | Prkcb | 4586 | 0.209 | 0.0852 | No |
| 33 | Ptprc | 4730 | 0.201 | 0.0831 | No |
| 34 | Flna | 4907 | 0.192 | 0.0790 | No |
| 35 | Prkcg | 4933 | 0.190 | 0.0828 | No |
| 36 | Ly75 | 5128 | 0.182 | 0.0775 | No |
| 37 | Traf2 | 5200 | 0.178 | 0.0785 | No |
| 38 | Fcgr2b | 5454 | 0.165 | 0.0698 | No |
| 39 | St8sia4 | 5517 | 0.161 | 0.0708 | No |
| 40 | Ccr5 | 5771 | 0.150 | 0.0617 | No |
| 41 | Icosl | 5898 | 0.144 | 0.0590 | No |
| 42 | Cd80 | 5981 | 0.139 | 0.0584 | No |
| 43 | Thy1 | 6023 | 0.137 | 0.0599 | No |
| 44 | Abi1 | 6047 | 0.136 | 0.0623 | No |
| 45 | Hcls1 | 6293 | 0.125 | 0.0529 | No |
| 46 | Nlrp3 | 6422 | 0.121 | 0.0495 | No |
| 47 | Eif3j1 | 6451 | 0.120 | 0.0512 | No |
| 48 | Nck1 | 6480 | 0.119 | 0.0529 | No |
| 49 | Map3k7 | 6567 | 0.115 | 0.0515 | No |
| 50 | Wars1 | 6684 | 0.109 | 0.0484 | No |
| 51 | Acvr2a | 6755 | 0.106 | 0.0475 | No |
| 52 | Degs1 | 6829 | 0.102 | 0.0464 | No |
| 53 | Irf4 | 7412 | 0.078 | 0.0184 | No |
| 54 | Il2 | 7561 | 0.073 | 0.0127 | No |
| 55 | Socs5 | 7873 | 0.060 | -0.0018 | No |
| 56 | Il7 | 8038 | 0.054 | -0.0088 | No |
| 57 | Srgn | 8545 | 0.033 | -0.0341 | No |
| 58 | Jak2 | 8599 | 0.030 | -0.0360 | No |
| 59 | Cd74 | 8661 | 0.028 | -0.0384 | No |
| 60 | Hif1a | 9076 | 0.013 | -0.0595 | No |
| 61 | Cd47 | 9118 | 0.011 | -0.0613 | No |
| 62 | Eif4g3 | 9131 | 0.010 | -0.0617 | No |
| 63 | Gcnt1 | 9350 | 0.003 | -0.0728 | No |
| 64 | Ly86 | 9611 | -0.005 | -0.0861 | No |
| 65 | Il16 | 10309 | -0.033 | -0.1213 | No |
| 66 | Ncf4 | 10624 | -0.045 | -0.1363 | No |
| 67 | Npm1 | 10671 | -0.046 | -0.1374 | No |
| 68 | Mrpl3 | 10689 | -0.047 | -0.1371 | No |
| 69 | Ifngr1 | 10873 | -0.055 | -0.1451 | No |
| 70 | Apbb1 | 10990 | -0.060 | -0.1495 | No |
| 71 | Il12a | 11150 | -0.066 | -0.1560 | No |
| 72 | Csf1 | 11155 | -0.066 | -0.1545 | No |
| 73 | Ikbkb | 11220 | -0.068 | -0.1560 | No |
| 74 | Igsf6 | 11327 | -0.072 | -0.1595 | No |
| 75 | Mtif2 | 11358 | -0.074 | -0.1591 | No |
| 76 | Cd247 | 11366 | -0.074 | -0.1576 | No |
| 77 | Spi1 | 11499 | -0.079 | -0.1623 | No |
| 78 | Map4k1 | 11747 | -0.091 | -0.1727 | No |
| 79 | Tpd52 | 11756 | -0.091 | -0.1707 | No |
| 80 | Ctss | 11800 | -0.093 | -0.1705 | No |
| 81 | Nme1 | 11923 | -0.098 | -0.1742 | No |
| 82 | Ccr2 | 11978 | -0.100 | -0.1743 | No |
| 83 | Was | 12098 | -0.106 | -0.1777 | No |
| 84 | Irf8 | 12238 | -0.112 | -0.1819 | No |
| 85 | Stat4 | 12463 | -0.121 | -0.1903 | No |
| 86 | Il11 | 12507 | -0.122 | -0.1893 | No |
| 87 | Csk | 12849 | -0.134 | -0.2034 | No |
| 88 | H2-Aa | 12911 | -0.137 | -0.2030 | No |
| 89 | Itgb2 | 13030 | -0.143 | -0.2053 | No |
| 90 | Lcp2 | 13096 | -0.145 | -0.2049 | No |
| 91 | Dyrk3 | 13348 | -0.158 | -0.2137 | No |
| 92 | Itgal | 13362 | -0.159 | -0.2102 | No |
| 93 | Glmn | 13461 | -0.163 | -0.2109 | No |
| 94 | Cxcr3 | 13463 | -0.163 | -0.2067 | No |
| 95 | Psmb10 | 13522 | -0.166 | -0.2053 | No |
| 96 | Eif3d | 13739 | -0.175 | -0.2119 | No |
| 97 | Tapbp | 13775 | -0.177 | -0.2090 | No |
| 98 | Ccnd3 | 13876 | -0.181 | -0.2095 | No |
| 99 | Ptpn6 | 14039 | -0.189 | -0.2129 | No |
| 100 | Il2rg | 14181 | -0.195 | -0.2150 | No |
| 101 | Hdac9 | 14244 | -0.199 | -0.2130 | No |
| 102 | Ripk2 | 14426 | -0.207 | -0.2169 | No |
| 103 | Tnf | 14791 | -0.225 | -0.2298 | No |
| 104 | Lyn | 14874 | -0.230 | -0.2280 | No |
| 105 | Cfp | 14938 | -0.233 | -0.2251 | No |
| 106 | Trat1 | 15060 | -0.240 | -0.2250 | No |
| 107 | Ache | 15206 | -0.247 | -0.2260 | No |
| 108 | Cd96 | 15219 | -0.248 | -0.2201 | No |
| 109 | Tlr1 | 15667 | -0.273 | -0.2360 | No |
| 110 | Eif5a | 15762 | -0.279 | -0.2336 | No |
| 111 | Pf4 | 16019 | -0.294 | -0.2391 | Yes |
| 112 | Rps9 | 16033 | -0.295 | -0.2320 | Yes |
| 113 | Stab1 | 16160 | -0.301 | -0.2306 | Yes |
| 114 | Cd40 | 16177 | -0.301 | -0.2235 | Yes |
| 115 | Irf7 | 16223 | -0.303 | -0.2178 | Yes |
| 116 | Tap2 | 16244 | -0.305 | -0.2108 | Yes |
| 117 | Bcl10 | 16270 | -0.307 | -0.2041 | Yes |
| 118 | Tgfb1 | 16282 | -0.307 | -0.1966 | Yes |
| 119 | Ccl5 | 16319 | -0.310 | -0.1903 | Yes |
| 120 | Tlr6 | 16370 | -0.313 | -0.1846 | Yes |
| 121 | Rpl9 | 16536 | -0.322 | -0.1847 | Yes |
| 122 | Il15 | 16988 | -0.354 | -0.1987 | Yes |
| 123 | Ltb | 17091 | -0.361 | -0.1945 | Yes |
| 124 | Rps3a1 | 17289 | -0.375 | -0.1948 | Yes |
| 125 | Lck | 17337 | -0.379 | -0.1872 | Yes |
| 126 | Tap1 | 17537 | -0.397 | -0.1871 | Yes |
| 127 | Il18 | 17710 | -0.412 | -0.1851 | Yes |
| 128 | F2r | 17793 | -0.420 | -0.1783 | Yes |
| 129 | Ifnar2 | 17964 | -0.438 | -0.1756 | Yes |
| 130 | H2-DMa | 17975 | -0.440 | -0.1646 | Yes |
| 131 | Capg | 17995 | -0.442 | -0.1539 | Yes |
| 132 | Il12rb1 | 18067 | -0.449 | -0.1458 | Yes |
| 133 | Tlr3 | 18100 | -0.453 | -0.1355 | Yes |
| 134 | B2m | 18217 | -0.466 | -0.1293 | Yes |
| 135 | C2 | 18243 | -0.470 | -0.1182 | Yes |
| 136 | Ube2d1 | 18295 | -0.476 | -0.1084 | Yes |
| 137 | Bcat1 | 18417 | -0.496 | -0.1016 | Yes |
| 138 | Socs1 | 18462 | -0.502 | -0.0907 | Yes |
| 139 | Elf4 | 18545 | -0.515 | -0.0814 | Yes |
| 140 | H2-Oa | 18847 | -0.570 | -0.0819 | Yes |
| 141 | Ccnd2 | 18993 | -0.609 | -0.0734 | Yes |
| 142 | Cd1d2 | 19015 | -0.616 | -0.0583 | Yes |
| 143 | H2-DMb2 | 19050 | -0.630 | -0.0435 | Yes |
| 144 | Cd86 | 19275 | -0.741 | -0.0356 | Yes |
| 145 | Rpl3l | 19421 | -0.872 | -0.0202 | Yes |
| 146 | H2-Ob | 19469 | -0.944 | 0.0022 | Yes |