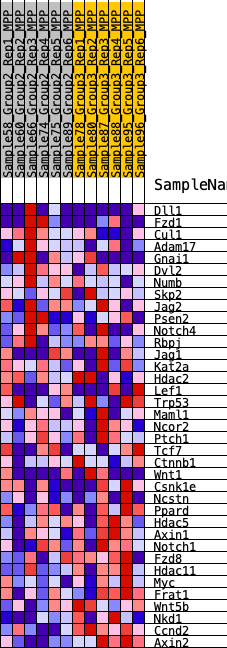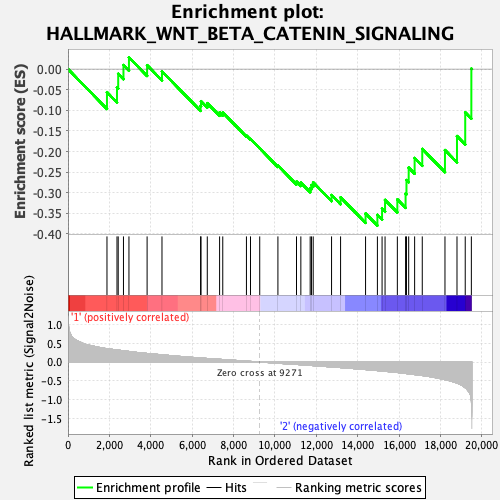
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.3798015 |
| Normalized Enrichment Score (NES) | -1.2680861 |
| Nominal p-value | 0.21471173 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.871 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dll1 | 1883 | 0.357 | -0.0561 | No |
| 2 | Fzd1 | 2367 | 0.320 | -0.0445 | No |
| 3 | Cul1 | 2426 | 0.316 | -0.0114 | No |
| 4 | Adam17 | 2680 | 0.300 | 0.0098 | No |
| 5 | Gnai1 | 2945 | 0.282 | 0.0284 | No |
| 6 | Dvl2 | 3822 | 0.228 | 0.0094 | No |
| 7 | Numb | 4541 | 0.192 | -0.0056 | No |
| 8 | Skp2 | 6406 | 0.107 | -0.0891 | No |
| 9 | Jag2 | 6427 | 0.107 | -0.0780 | No |
| 10 | Psen2 | 6730 | 0.094 | -0.0828 | No |
| 11 | Notch4 | 7325 | 0.072 | -0.1051 | No |
| 12 | Rbpj | 7478 | 0.067 | -0.1053 | No |
| 13 | Jag1 | 8622 | 0.023 | -0.1614 | No |
| 14 | Kat2a | 8818 | 0.016 | -0.1696 | No |
| 15 | Hdac2 | 9264 | 0.000 | -0.1924 | No |
| 16 | Lef1 | 10143 | -0.030 | -0.2340 | No |
| 17 | Trp53 | 11041 | -0.063 | -0.2729 | No |
| 18 | Maml1 | 11251 | -0.071 | -0.2756 | No |
| 19 | Ncor2 | 11702 | -0.088 | -0.2887 | No |
| 20 | Ptch1 | 11764 | -0.091 | -0.2815 | No |
| 21 | Tcf7 | 11848 | -0.094 | -0.2750 | No |
| 22 | Ctnnb1 | 12735 | -0.130 | -0.3057 | No |
| 23 | Wnt1 | 13171 | -0.147 | -0.3113 | No |
| 24 | Csnk1e | 14378 | -0.199 | -0.3505 | No |
| 25 | Ncstn | 14950 | -0.227 | -0.3539 | Yes |
| 26 | Ppard | 15176 | -0.238 | -0.3383 | Yes |
| 27 | Hdac5 | 15320 | -0.247 | -0.3175 | Yes |
| 28 | Axin1 | 15914 | -0.278 | -0.3164 | Yes |
| 29 | Notch1 | 16312 | -0.304 | -0.3021 | Yes |
| 30 | Fzd8 | 16355 | -0.307 | -0.2693 | Yes |
| 31 | Hdac11 | 16462 | -0.313 | -0.2391 | Yes |
| 32 | Myc | 16749 | -0.334 | -0.2158 | Yes |
| 33 | Frat1 | 17118 | -0.359 | -0.1938 | Yes |
| 34 | Wnt5b | 18215 | -0.470 | -0.1966 | Yes |
| 35 | Nkd1 | 18796 | -0.560 | -0.1626 | Yes |
| 36 | Ccnd2 | 19194 | -0.684 | -0.1051 | Yes |
| 37 | Axin2 | 19490 | -1.066 | 0.0011 | Yes |