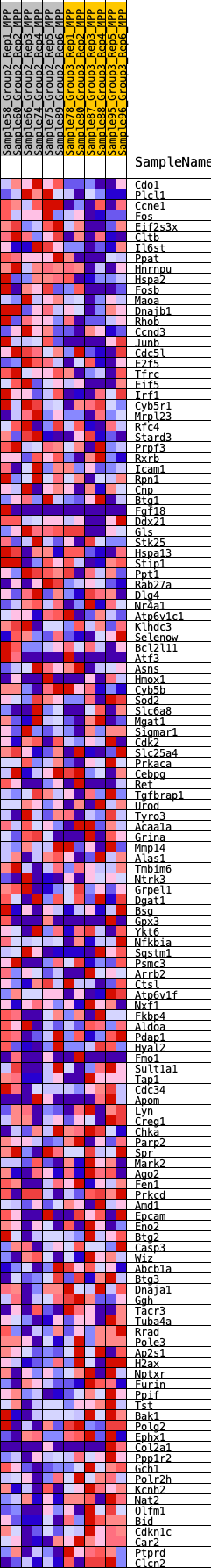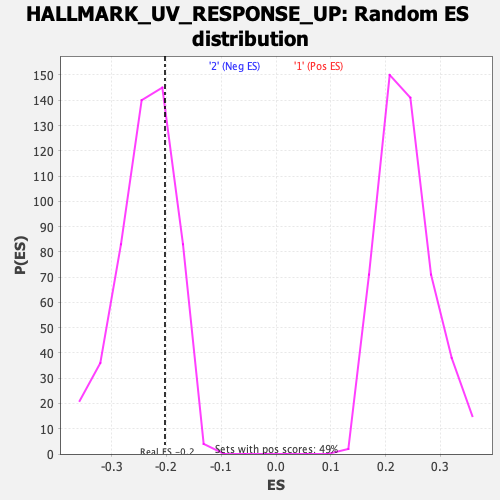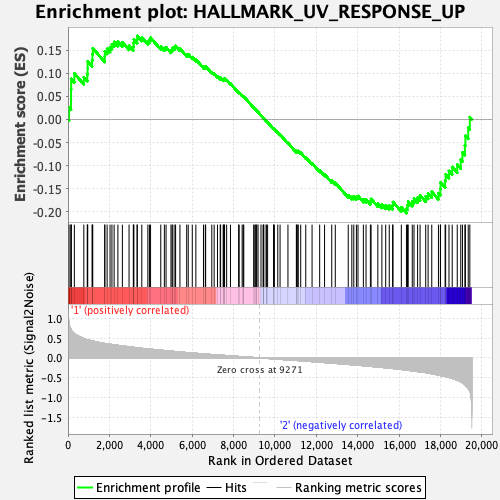
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.20255612 |
| Normalized Enrichment Score (NES) | -0.85640025 |
| Nominal p-value | 0.7207031 |
| FDR q-value | 0.97388256 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdo1 | 51 | 0.885 | 0.0255 | No |
| 2 | Plcl1 | 146 | 0.712 | 0.0433 | No |
| 3 | Ccne1 | 148 | 0.708 | 0.0657 | No |
| 4 | Fos | 154 | 0.699 | 0.0877 | No |
| 5 | Eif2s3x | 307 | 0.613 | 0.0993 | No |
| 6 | Cltb | 765 | 0.487 | 0.0912 | No |
| 7 | Il6st | 930 | 0.461 | 0.0974 | No |
| 8 | Ppat | 948 | 0.457 | 0.1111 | No |
| 9 | Hnrnpu | 949 | 0.457 | 0.1256 | No |
| 10 | Hspa2 | 1163 | 0.430 | 0.1283 | No |
| 11 | Fosb | 1183 | 0.428 | 0.1409 | No |
| 12 | Maoa | 1196 | 0.426 | 0.1538 | No |
| 13 | Dnajb1 | 1775 | 0.365 | 0.1356 | No |
| 14 | Rhob | 1779 | 0.364 | 0.1470 | No |
| 15 | Ccnd3 | 1890 | 0.356 | 0.1527 | No |
| 16 | Junb | 2032 | 0.345 | 0.1563 | No |
| 17 | Cdc5l | 2123 | 0.339 | 0.1625 | No |
| 18 | E2f5 | 2229 | 0.330 | 0.1675 | No |
| 19 | Tfrc | 2411 | 0.318 | 0.1683 | No |
| 20 | Eif5 | 2627 | 0.303 | 0.1668 | No |
| 21 | Irf1 | 2950 | 0.282 | 0.1592 | No |
| 22 | Cyb5r1 | 3156 | 0.268 | 0.1571 | No |
| 23 | Mrpl23 | 3169 | 0.267 | 0.1650 | No |
| 24 | Rfc4 | 3181 | 0.266 | 0.1728 | No |
| 25 | Stard3 | 3339 | 0.255 | 0.1728 | No |
| 26 | Prpf3 | 3347 | 0.254 | 0.1806 | No |
| 27 | Rxrb | 3566 | 0.242 | 0.1770 | No |
| 28 | Icam1 | 3859 | 0.226 | 0.1691 | No |
| 29 | Rpn1 | 3935 | 0.222 | 0.1723 | No |
| 30 | Cnp | 3989 | 0.220 | 0.1766 | No |
| 31 | Btg1 | 4483 | 0.195 | 0.1574 | No |
| 32 | Fgf18 | 4656 | 0.187 | 0.1544 | No |
| 33 | Ddx21 | 4738 | 0.183 | 0.1561 | No |
| 34 | Gls | 4976 | 0.172 | 0.1493 | No |
| 35 | Stk25 | 5030 | 0.169 | 0.1519 | No |
| 36 | Hspa13 | 5067 | 0.167 | 0.1554 | No |
| 37 | Stip1 | 5167 | 0.162 | 0.1554 | No |
| 38 | Ppt1 | 5199 | 0.161 | 0.1589 | No |
| 39 | Rab27a | 5408 | 0.150 | 0.1529 | No |
| 40 | Dlg4 | 5735 | 0.135 | 0.1404 | No |
| 41 | Nr4a1 | 5810 | 0.132 | 0.1408 | No |
| 42 | Atp6v1c1 | 6005 | 0.124 | 0.1347 | No |
| 43 | Klhdc3 | 6178 | 0.118 | 0.1296 | No |
| 44 | Selenow | 6548 | 0.101 | 0.1138 | No |
| 45 | Bcl2l11 | 6635 | 0.098 | 0.1124 | No |
| 46 | Atf3 | 6651 | 0.098 | 0.1148 | No |
| 47 | Asns | 6948 | 0.086 | 0.1022 | No |
| 48 | Hmox1 | 7062 | 0.081 | 0.0990 | No |
| 49 | Cyb5b | 7223 | 0.076 | 0.0931 | No |
| 50 | Sod2 | 7355 | 0.071 | 0.0886 | No |
| 51 | Slc6a8 | 7374 | 0.070 | 0.0899 | No |
| 52 | Mgat1 | 7502 | 0.066 | 0.0854 | No |
| 53 | Sigmar1 | 7508 | 0.065 | 0.0873 | No |
| 54 | Cdk2 | 7557 | 0.064 | 0.0868 | No |
| 55 | Slc25a4 | 7566 | 0.063 | 0.0884 | No |
| 56 | Prkaca | 7666 | 0.060 | 0.0852 | No |
| 57 | Cebpg | 7850 | 0.053 | 0.0774 | No |
| 58 | Ret | 8245 | 0.037 | 0.0583 | No |
| 59 | Tgfbrap1 | 8270 | 0.036 | 0.0582 | No |
| 60 | Urod | 8415 | 0.031 | 0.0518 | No |
| 61 | Tyro3 | 8468 | 0.029 | 0.0500 | No |
| 62 | Acaa1a | 8493 | 0.028 | 0.0497 | No |
| 63 | Grina | 8970 | 0.012 | 0.0255 | No |
| 64 | Mmp14 | 9029 | 0.009 | 0.0228 | No |
| 65 | Alas1 | 9049 | 0.008 | 0.0221 | No |
| 66 | Tmbim6 | 9098 | 0.007 | 0.0198 | No |
| 67 | Ntrk3 | 9119 | 0.006 | 0.0190 | No |
| 68 | Grpel1 | 9183 | 0.004 | 0.0158 | No |
| 69 | Dgat1 | 9325 | -0.001 | 0.0086 | No |
| 70 | Bsg | 9427 | -0.004 | 0.0035 | No |
| 71 | Gpx3 | 9444 | -0.005 | 0.0028 | No |
| 72 | Ykt6 | 9463 | -0.006 | 0.0021 | No |
| 73 | Nfkbia | 9578 | -0.010 | -0.0035 | No |
| 74 | Sqstm1 | 9579 | -0.010 | -0.0032 | No |
| 75 | Psmc3 | 9645 | -0.012 | -0.0061 | No |
| 76 | Arrb2 | 9934 | -0.022 | -0.0203 | No |
| 77 | Ctsl | 9939 | -0.023 | -0.0198 | No |
| 78 | Atp6v1f | 9971 | -0.024 | -0.0206 | No |
| 79 | Nxf1 | 10136 | -0.030 | -0.0281 | No |
| 80 | Fkbp4 | 10247 | -0.034 | -0.0327 | No |
| 81 | Aldoa | 10628 | -0.048 | -0.0508 | No |
| 82 | Pdap1 | 11034 | -0.062 | -0.0697 | No |
| 83 | Hyal2 | 11044 | -0.063 | -0.0682 | No |
| 84 | Fmo1 | 11099 | -0.065 | -0.0689 | No |
| 85 | Sult1a1 | 11124 | -0.066 | -0.0680 | No |
| 86 | Tap1 | 11244 | -0.071 | -0.0719 | No |
| 87 | Cdc34 | 11488 | -0.079 | -0.0819 | No |
| 88 | Apom | 11793 | -0.092 | -0.0947 | No |
| 89 | Lyn | 12160 | -0.106 | -0.1102 | No |
| 90 | Creg1 | 12397 | -0.116 | -0.1187 | No |
| 91 | Chka | 12744 | -0.130 | -0.1324 | No |
| 92 | Parp2 | 12915 | -0.137 | -0.1368 | No |
| 93 | Spr | 13541 | -0.163 | -0.1639 | No |
| 94 | Mark2 | 13714 | -0.171 | -0.1673 | No |
| 95 | Ago2 | 13809 | -0.174 | -0.1666 | No |
| 96 | Fen1 | 13936 | -0.180 | -0.1674 | No |
| 97 | Prkcd | 14012 | -0.183 | -0.1655 | No |
| 98 | Amd1 | 14272 | -0.195 | -0.1726 | No |
| 99 | Epcam | 14401 | -0.201 | -0.1728 | No |
| 100 | Eno2 | 14611 | -0.211 | -0.1769 | No |
| 101 | Btg2 | 14644 | -0.213 | -0.1718 | No |
| 102 | Casp3 | 14971 | -0.228 | -0.1814 | No |
| 103 | Wiz | 15167 | -0.238 | -0.1839 | No |
| 104 | Abcb1a | 15351 | -0.248 | -0.1854 | No |
| 105 | Btg3 | 15523 | -0.257 | -0.1861 | No |
| 106 | Dnaja1 | 15684 | -0.266 | -0.1859 | No |
| 107 | Ggh | 15704 | -0.267 | -0.1784 | No |
| 108 | Tacr3 | 16106 | -0.290 | -0.1899 | No |
| 109 | Tuba4a | 16353 | -0.307 | -0.1928 | Yes |
| 110 | Rrad | 16404 | -0.310 | -0.1855 | Yes |
| 111 | Pole3 | 16438 | -0.312 | -0.1773 | Yes |
| 112 | Ap2s1 | 16635 | -0.325 | -0.1771 | Yes |
| 113 | H2ax | 16719 | -0.331 | -0.1709 | Yes |
| 114 | Nptxr | 16893 | -0.343 | -0.1689 | Yes |
| 115 | Furin | 17012 | -0.351 | -0.1638 | Yes |
| 116 | Ppif | 17284 | -0.370 | -0.1660 | Yes |
| 117 | Tst | 17400 | -0.380 | -0.1599 | Yes |
| 118 | Bak1 | 17577 | -0.399 | -0.1563 | Yes |
| 119 | Polg2 | 17900 | -0.432 | -0.1592 | Yes |
| 120 | Ephx1 | 17993 | -0.443 | -0.1498 | Yes |
| 121 | Col2a1 | 18001 | -0.444 | -0.1361 | Yes |
| 122 | Ppp1r2 | 18222 | -0.470 | -0.1325 | Yes |
| 123 | Gch1 | 18248 | -0.474 | -0.1187 | Yes |
| 124 | Polr2h | 18409 | -0.494 | -0.1113 | Yes |
| 125 | Kcnh2 | 18565 | -0.518 | -0.1028 | Yes |
| 126 | Nat2 | 18808 | -0.564 | -0.0974 | Yes |
| 127 | Olfm1 | 18978 | -0.603 | -0.0870 | Yes |
| 128 | Bid | 19060 | -0.626 | -0.0712 | Yes |
| 129 | Cdkn1c | 19179 | -0.679 | -0.0558 | Yes |
| 130 | Car2 | 19202 | -0.687 | -0.0351 | Yes |
| 131 | Ptprd | 19339 | -0.768 | -0.0177 | Yes |
| 132 | Clcn2 | 19416 | -0.835 | 0.0050 | Yes |