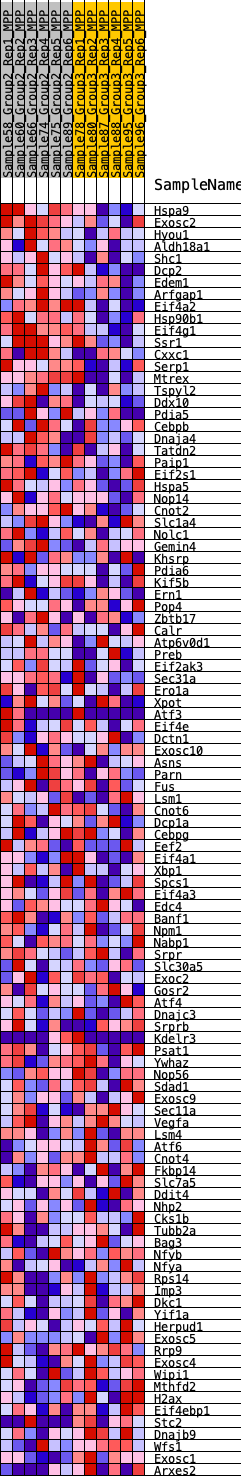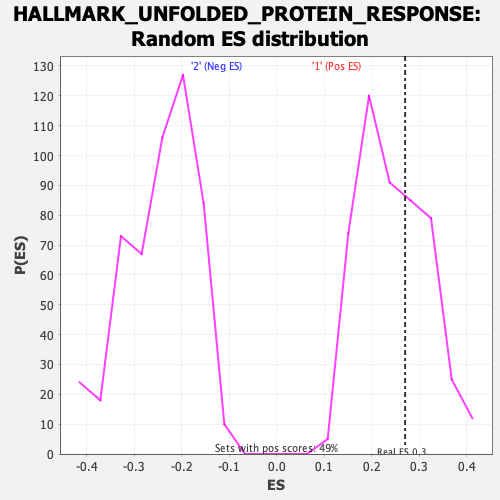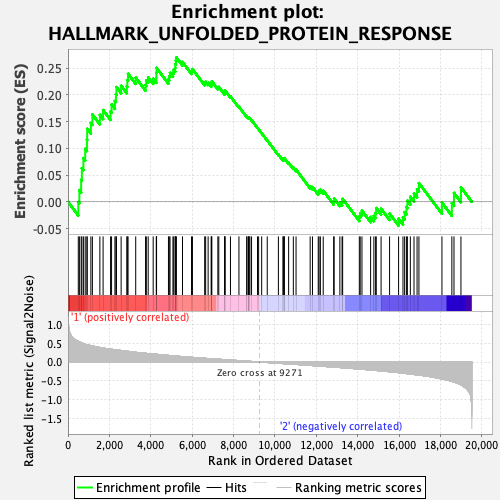
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.27007973 |
| Normalized Enrichment Score (NES) | 1.0981957 |
| Nominal p-value | 0.36863545 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.981 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hspa9 | 497 | 0.554 | -0.0002 | Yes |
| 2 | Exosc2 | 550 | 0.540 | 0.0218 | Yes |
| 3 | Hyou1 | 636 | 0.520 | 0.0413 | Yes |
| 4 | Aldh18a1 | 670 | 0.510 | 0.0629 | Yes |
| 5 | Shc1 | 744 | 0.492 | 0.0817 | Yes |
| 6 | Dcp2 | 831 | 0.476 | 0.0991 | Yes |
| 7 | Edem1 | 915 | 0.464 | 0.1160 | Yes |
| 8 | Arfgap1 | 927 | 0.461 | 0.1366 | Yes |
| 9 | Eif4a2 | 1105 | 0.438 | 0.1475 | Yes |
| 10 | Hsp90b1 | 1178 | 0.429 | 0.1635 | Yes |
| 11 | Eif4g1 | 1540 | 0.388 | 0.1626 | Yes |
| 12 | Ssr1 | 1696 | 0.372 | 0.1717 | Yes |
| 13 | Cxxc1 | 2057 | 0.344 | 0.1689 | Yes |
| 14 | Serp1 | 2105 | 0.340 | 0.1820 | Yes |
| 15 | Mtrex | 2267 | 0.327 | 0.1887 | Yes |
| 16 | Tspyl2 | 2316 | 0.324 | 0.2011 | Yes |
| 17 | Ddx10 | 2342 | 0.322 | 0.2146 | Yes |
| 18 | Pdia5 | 2567 | 0.307 | 0.2171 | Yes |
| 19 | Cebpb | 2843 | 0.288 | 0.2161 | Yes |
| 20 | Dnaja4 | 2866 | 0.287 | 0.2282 | Yes |
| 21 | Tatdn2 | 2902 | 0.285 | 0.2394 | Yes |
| 22 | Paip1 | 3272 | 0.260 | 0.2323 | Yes |
| 23 | Eif2s1 | 3747 | 0.232 | 0.2185 | Yes |
| 24 | Hspa5 | 3784 | 0.230 | 0.2272 | Yes |
| 25 | Nop14 | 3874 | 0.226 | 0.2329 | Yes |
| 26 | Cnot2 | 4116 | 0.215 | 0.2304 | Yes |
| 27 | Slc1a4 | 4270 | 0.206 | 0.2319 | Yes |
| 28 | Nolc1 | 4271 | 0.206 | 0.2414 | Yes |
| 29 | Gemin4 | 4279 | 0.206 | 0.2504 | Yes |
| 30 | Khsrp | 4854 | 0.177 | 0.2289 | Yes |
| 31 | Pdia6 | 4890 | 0.175 | 0.2351 | Yes |
| 32 | Kif5b | 4929 | 0.173 | 0.2411 | Yes |
| 33 | Ern1 | 5071 | 0.167 | 0.2415 | Yes |
| 34 | Pop4 | 5110 | 0.165 | 0.2471 | Yes |
| 35 | Zbtb17 | 5182 | 0.161 | 0.2508 | Yes |
| 36 | Calr | 5186 | 0.161 | 0.2580 | Yes |
| 37 | Atp6v0d1 | 5213 | 0.160 | 0.2640 | Yes |
| 38 | Preb | 5237 | 0.158 | 0.2701 | Yes |
| 39 | Eif2ak3 | 5532 | 0.144 | 0.2615 | No |
| 40 | Sec31a | 5969 | 0.125 | 0.2448 | No |
| 41 | Ero1a | 6013 | 0.123 | 0.2482 | No |
| 42 | Xpot | 6604 | 0.099 | 0.2224 | No |
| 43 | Atf3 | 6651 | 0.098 | 0.2245 | No |
| 44 | Eif4e | 6771 | 0.092 | 0.2226 | No |
| 45 | Dctn1 | 6917 | 0.087 | 0.2190 | No |
| 46 | Exosc10 | 6937 | 0.086 | 0.2220 | No |
| 47 | Asns | 6948 | 0.086 | 0.2254 | No |
| 48 | Parn | 7241 | 0.075 | 0.2138 | No |
| 49 | Fus | 7286 | 0.073 | 0.2149 | No |
| 50 | Lsm1 | 7567 | 0.063 | 0.2034 | No |
| 51 | Cnot6 | 7580 | 0.063 | 0.2056 | No |
| 52 | Dcp1a | 7588 | 0.063 | 0.2081 | No |
| 53 | Cebpg | 7850 | 0.053 | 0.1971 | No |
| 54 | Eef2 | 8260 | 0.037 | 0.1777 | No |
| 55 | Eif4a1 | 8636 | 0.022 | 0.1594 | No |
| 56 | Xbp1 | 8648 | 0.022 | 0.1599 | No |
| 57 | Spcs1 | 8721 | 0.019 | 0.1570 | No |
| 58 | Eif4a3 | 8742 | 0.019 | 0.1568 | No |
| 59 | Edc4 | 8744 | 0.018 | 0.1576 | No |
| 60 | Banf1 | 8789 | 0.017 | 0.1561 | No |
| 61 | Npm1 | 8869 | 0.015 | 0.1528 | No |
| 62 | Nabp1 | 9157 | 0.005 | 0.1382 | No |
| 63 | Srpr | 9211 | 0.002 | 0.1355 | No |
| 64 | Slc30a5 | 9358 | -0.002 | 0.1281 | No |
| 65 | Exoc2 | 9627 | -0.012 | 0.1148 | No |
| 66 | Gosr2 | 10166 | -0.031 | 0.0885 | No |
| 67 | Atf4 | 10386 | -0.040 | 0.0791 | No |
| 68 | Dnajc3 | 10408 | -0.041 | 0.0799 | No |
| 69 | Srprb | 10449 | -0.042 | 0.0797 | No |
| 70 | Kdelr3 | 10450 | -0.042 | 0.0816 | No |
| 71 | Psat1 | 10661 | -0.049 | 0.0731 | No |
| 72 | Ywhaz | 10888 | -0.058 | 0.0641 | No |
| 73 | Nop56 | 11019 | -0.062 | 0.0602 | No |
| 74 | Sdad1 | 11701 | -0.088 | 0.0292 | No |
| 75 | Exosc9 | 11813 | -0.093 | 0.0277 | No |
| 76 | Sec11a | 12091 | -0.103 | 0.0182 | No |
| 77 | Vegfa | 12131 | -0.105 | 0.0210 | No |
| 78 | Lsm4 | 12195 | -0.108 | 0.0226 | No |
| 79 | Atf6 | 12332 | -0.113 | 0.0208 | No |
| 80 | Cnot4 | 12837 | -0.134 | 0.0010 | No |
| 81 | Fkbp14 | 12859 | -0.135 | 0.0061 | No |
| 82 | Slc7a5 | 13136 | -0.146 | -0.0015 | No |
| 83 | Ddit4 | 13242 | -0.149 | -0.0000 | No |
| 84 | Nhp2 | 13271 | -0.151 | 0.0054 | No |
| 85 | Cks1b | 14076 | -0.186 | -0.0275 | No |
| 86 | Tubb2a | 14119 | -0.188 | -0.0210 | No |
| 87 | Bag3 | 14204 | -0.192 | -0.0166 | No |
| 88 | Nfyb | 14622 | -0.212 | -0.0284 | No |
| 89 | Nfya | 14779 | -0.219 | -0.0264 | No |
| 90 | Rps14 | 14864 | -0.223 | -0.0204 | No |
| 91 | Imp3 | 14894 | -0.225 | -0.0117 | No |
| 92 | Dkc1 | 15128 | -0.236 | -0.0128 | No |
| 93 | Yif1a | 15539 | -0.258 | -0.0222 | No |
| 94 | Herpud1 | 15974 | -0.281 | -0.0316 | No |
| 95 | Exosc5 | 16186 | -0.295 | -0.0290 | No |
| 96 | Rrp9 | 16266 | -0.301 | -0.0193 | No |
| 97 | Exosc4 | 16352 | -0.307 | -0.0096 | No |
| 98 | Wipi1 | 16399 | -0.310 | 0.0022 | No |
| 99 | Mthfd2 | 16542 | -0.319 | 0.0095 | No |
| 100 | H2ax | 16719 | -0.331 | 0.0156 | No |
| 101 | Eif4ebp1 | 16867 | -0.341 | 0.0237 | No |
| 102 | Stc2 | 16957 | -0.348 | 0.0350 | No |
| 103 | Dnajb9 | 18069 | -0.452 | -0.0015 | No |
| 104 | Wfs1 | 18548 | -0.515 | -0.0026 | No |
| 105 | Exosc1 | 18651 | -0.534 | 0.0166 | No |
| 106 | Arxes2 | 18987 | -0.605 | 0.0271 | No |