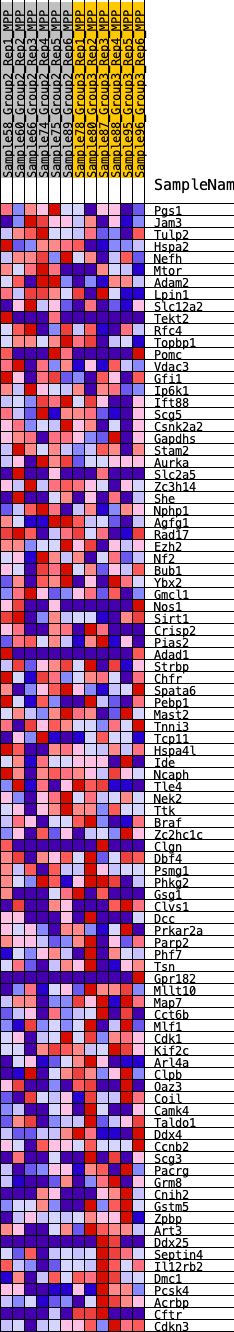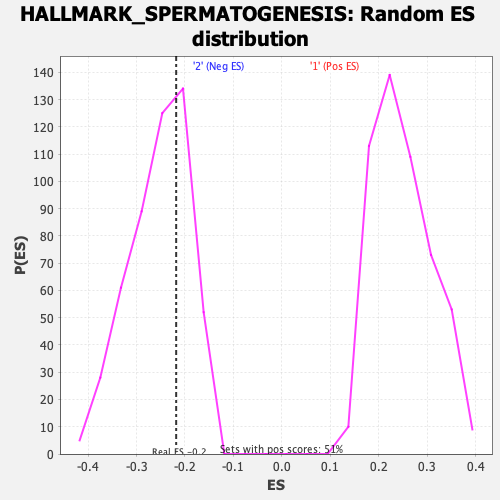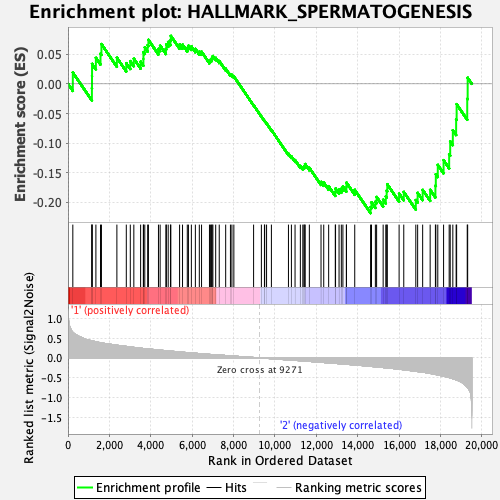
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.21809009 |
| Normalized Enrichment Score (NES) | -0.8602598 |
| Nominal p-value | 0.68218625 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pgs1 | 233 | 0.647 | 0.0192 | No |
| 2 | Jam3 | 1155 | 0.431 | -0.0075 | No |
| 3 | Tulp2 | 1160 | 0.431 | 0.0131 | No |
| 4 | Hspa2 | 1163 | 0.430 | 0.0337 | No |
| 5 | Nefh | 1348 | 0.408 | 0.0439 | No |
| 6 | Mtor | 1571 | 0.385 | 0.0511 | No |
| 7 | Adam2 | 1616 | 0.380 | 0.0671 | No |
| 8 | Lpin1 | 2362 | 0.320 | 0.0442 | No |
| 9 | Slc12a2 | 2817 | 0.291 | 0.0349 | No |
| 10 | Tekt2 | 3010 | 0.278 | 0.0384 | No |
| 11 | Rfc4 | 3181 | 0.266 | 0.0424 | No |
| 12 | Topbp1 | 3509 | 0.245 | 0.0374 | No |
| 13 | Pomc | 3643 | 0.238 | 0.0421 | No |
| 14 | Vdac3 | 3645 | 0.238 | 0.0535 | No |
| 15 | Gfi1 | 3710 | 0.235 | 0.0615 | No |
| 16 | Ip6k1 | 3850 | 0.227 | 0.0653 | No |
| 17 | Ift88 | 3886 | 0.225 | 0.0743 | No |
| 18 | Scg5 | 4371 | 0.201 | 0.0591 | No |
| 19 | Csnk2a2 | 4452 | 0.198 | 0.0645 | No |
| 20 | Gapdhs | 4731 | 0.183 | 0.0590 | No |
| 21 | Stam2 | 4753 | 0.183 | 0.0667 | No |
| 22 | Aurka | 4841 | 0.177 | 0.0708 | No |
| 23 | Slc2a5 | 4951 | 0.173 | 0.0735 | No |
| 24 | Zc3h14 | 4969 | 0.172 | 0.0809 | No |
| 25 | She | 5392 | 0.150 | 0.0665 | No |
| 26 | Nphp1 | 5534 | 0.144 | 0.0661 | No |
| 27 | Agfg1 | 5764 | 0.134 | 0.0608 | No |
| 28 | Rad17 | 5819 | 0.132 | 0.0644 | No |
| 29 | Ezh2 | 5961 | 0.126 | 0.0632 | No |
| 30 | Nf2 | 6154 | 0.119 | 0.0590 | No |
| 31 | Bub1 | 6345 | 0.110 | 0.0546 | No |
| 32 | Ybx2 | 6453 | 0.105 | 0.0541 | No |
| 33 | Gmcl1 | 6835 | 0.090 | 0.0388 | No |
| 34 | Nos1 | 6881 | 0.088 | 0.0408 | No |
| 35 | Sirt1 | 6938 | 0.086 | 0.0420 | No |
| 36 | Crisp2 | 6964 | 0.085 | 0.0448 | No |
| 37 | Pias2 | 7001 | 0.083 | 0.0470 | No |
| 38 | Adad1 | 7136 | 0.079 | 0.0439 | No |
| 39 | Strbp | 7308 | 0.073 | 0.0386 | No |
| 40 | Chfr | 7619 | 0.062 | 0.0256 | No |
| 41 | Spata6 | 7849 | 0.053 | 0.0163 | No |
| 42 | Pebp1 | 7920 | 0.050 | 0.0151 | No |
| 43 | Mast2 | 8014 | 0.047 | 0.0126 | No |
| 44 | Tnni3 | 8968 | 0.012 | -0.0359 | No |
| 45 | Tcp11 | 9344 | -0.002 | -0.0552 | No |
| 46 | Hspa4l | 9492 | -0.007 | -0.0624 | No |
| 47 | Ide | 9504 | -0.007 | -0.0626 | No |
| 48 | Ncaph | 9595 | -0.011 | -0.0667 | No |
| 49 | Tle4 | 9828 | -0.019 | -0.0778 | No |
| 50 | Nek2 | 10652 | -0.049 | -0.1178 | No |
| 51 | Ttk | 10795 | -0.054 | -0.1225 | No |
| 52 | Braf | 10971 | -0.060 | -0.1286 | No |
| 53 | Zc2hc1c | 11227 | -0.070 | -0.1384 | No |
| 54 | Clgn | 11347 | -0.074 | -0.1409 | No |
| 55 | Dbf4 | 11399 | -0.076 | -0.1399 | No |
| 56 | Psmg1 | 11439 | -0.078 | -0.1381 | No |
| 57 | Phkg2 | 11457 | -0.079 | -0.1352 | No |
| 58 | Gsg1 | 11662 | -0.086 | -0.1416 | No |
| 59 | Clvs1 | 12226 | -0.109 | -0.1653 | No |
| 60 | Dcc | 12357 | -0.115 | -0.1665 | No |
| 61 | Prkar2a | 12595 | -0.124 | -0.1727 | No |
| 62 | Parp2 | 12915 | -0.137 | -0.1825 | No |
| 63 | Phf7 | 12928 | -0.138 | -0.1765 | No |
| 64 | Tsn | 13097 | -0.144 | -0.1782 | No |
| 65 | Gpr182 | 13207 | -0.148 | -0.1766 | No |
| 66 | Mllt10 | 13283 | -0.151 | -0.1732 | No |
| 67 | Map7 | 13452 | -0.159 | -0.1742 | No |
| 68 | Cct6b | 13454 | -0.159 | -0.1666 | No |
| 69 | Mlf1 | 13855 | -0.176 | -0.1787 | No |
| 70 | Cdk1 | 14621 | -0.212 | -0.2079 | Yes |
| 71 | Kif2c | 14665 | -0.214 | -0.1998 | Yes |
| 72 | Arl4a | 14852 | -0.223 | -0.1986 | Yes |
| 73 | Clpb | 14912 | -0.225 | -0.1908 | Yes |
| 74 | Oaz3 | 15228 | -0.242 | -0.1953 | Yes |
| 75 | Coil | 15356 | -0.248 | -0.1899 | Yes |
| 76 | Camk4 | 15398 | -0.250 | -0.1800 | Yes |
| 77 | Taldo1 | 15428 | -0.252 | -0.1693 | Yes |
| 78 | Ddx4 | 15999 | -0.283 | -0.1850 | Yes |
| 79 | Ccnb2 | 16224 | -0.297 | -0.1822 | Yes |
| 80 | Scg3 | 16806 | -0.337 | -0.1959 | Yes |
| 81 | Pacrg | 16895 | -0.343 | -0.1839 | Yes |
| 82 | Grm8 | 17137 | -0.360 | -0.1789 | Yes |
| 83 | Cnih2 | 17499 | -0.390 | -0.1787 | Yes |
| 84 | Gstm5 | 17753 | -0.415 | -0.1717 | Yes |
| 85 | Zpbp | 17773 | -0.418 | -0.1525 | Yes |
| 86 | Art3 | 17868 | -0.429 | -0.1366 | Yes |
| 87 | Ddx25 | 18148 | -0.462 | -0.1287 | Yes |
| 88 | Septin4 | 18418 | -0.495 | -0.1187 | Yes |
| 89 | Il12rb2 | 18468 | -0.503 | -0.0970 | Yes |
| 90 | Dmc1 | 18595 | -0.523 | -0.0783 | Yes |
| 91 | Pcsk4 | 18755 | -0.551 | -0.0599 | Yes |
| 92 | Acrbp | 18779 | -0.555 | -0.0343 | Yes |
| 93 | Cftr | 19291 | -0.735 | -0.0251 | Yes |
| 94 | Cdkn3 | 19312 | -0.756 | 0.0103 | Yes |