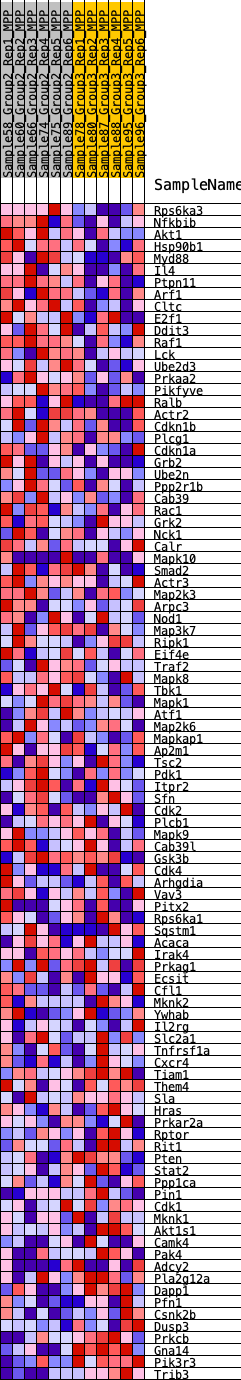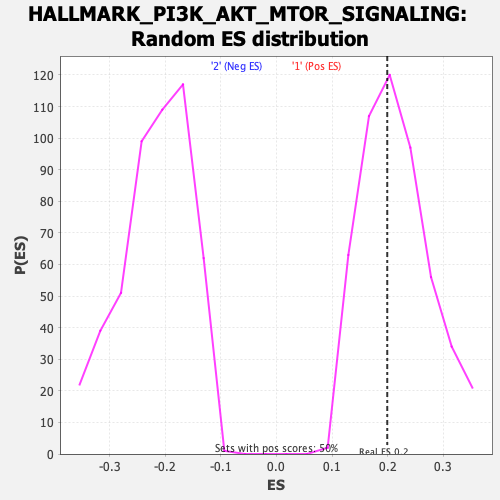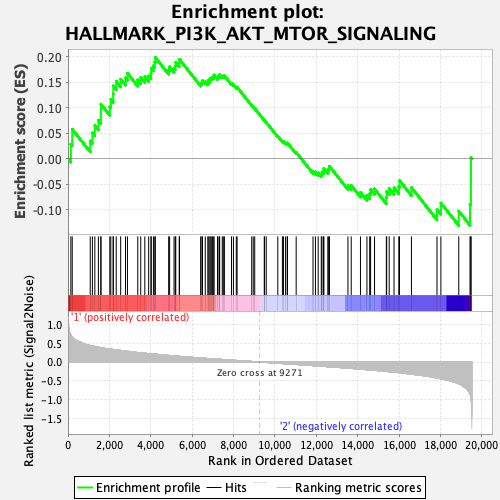
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.19906475 |
| Normalized Enrichment Score (NES) | 0.92166346 |
| Nominal p-value | 0.584 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rps6ka3 | 138 | 0.728 | 0.0287 | Yes |
| 2 | Nfkbib | 204 | 0.663 | 0.0580 | Yes |
| 3 | Akt1 | 1074 | 0.442 | 0.0350 | Yes |
| 4 | Hsp90b1 | 1178 | 0.429 | 0.0508 | Yes |
| 5 | Myd88 | 1297 | 0.414 | 0.0651 | Yes |
| 6 | Il4 | 1469 | 0.394 | 0.0757 | Yes |
| 7 | Ptpn11 | 1589 | 0.383 | 0.0884 | Yes |
| 8 | Arf1 | 1590 | 0.382 | 0.1072 | Yes |
| 9 | Cltc | 2021 | 0.346 | 0.1021 | Yes |
| 10 | E2f1 | 2068 | 0.343 | 0.1166 | Yes |
| 11 | Ddit3 | 2183 | 0.335 | 0.1272 | Yes |
| 12 | Raf1 | 2189 | 0.334 | 0.1434 | Yes |
| 13 | Lck | 2330 | 0.323 | 0.1522 | Yes |
| 14 | Ube2d3 | 2545 | 0.309 | 0.1563 | Yes |
| 15 | Prkaa2 | 2778 | 0.294 | 0.1588 | Yes |
| 16 | Pikfyve | 2872 | 0.286 | 0.1682 | Yes |
| 17 | Ralb | 3371 | 0.253 | 0.1550 | Yes |
| 18 | Actr2 | 3515 | 0.245 | 0.1597 | Yes |
| 19 | Cdkn1b | 3711 | 0.234 | 0.1612 | Yes |
| 20 | Plcg1 | 3897 | 0.224 | 0.1627 | Yes |
| 21 | Cdkn1a | 4008 | 0.219 | 0.1678 | Yes |
| 22 | Grb2 | 4021 | 0.219 | 0.1779 | Yes |
| 23 | Ube2n | 4128 | 0.214 | 0.1830 | Yes |
| 24 | Ppp2r1b | 4194 | 0.210 | 0.1900 | Yes |
| 25 | Cab39 | 4219 | 0.209 | 0.1991 | Yes |
| 26 | Rac1 | 4864 | 0.176 | 0.1746 | No |
| 27 | Grk2 | 4910 | 0.174 | 0.1808 | No |
| 28 | Nck1 | 5124 | 0.164 | 0.1779 | No |
| 29 | Calr | 5186 | 0.161 | 0.1827 | No |
| 30 | Mapk10 | 5207 | 0.160 | 0.1895 | No |
| 31 | Smad2 | 5375 | 0.151 | 0.1884 | No |
| 32 | Actr3 | 5386 | 0.151 | 0.1953 | No |
| 33 | Map2k3 | 6409 | 0.107 | 0.1479 | No |
| 34 | Arpc3 | 6470 | 0.105 | 0.1500 | No |
| 35 | Nod1 | 6500 | 0.103 | 0.1536 | No |
| 36 | Map3k7 | 6633 | 0.098 | 0.1516 | No |
| 37 | Ripk1 | 6765 | 0.092 | 0.1494 | No |
| 38 | Eif4e | 6771 | 0.092 | 0.1537 | No |
| 39 | Traf2 | 6861 | 0.089 | 0.1535 | No |
| 40 | Mapk8 | 6862 | 0.089 | 0.1578 | No |
| 41 | Tbk1 | 6923 | 0.086 | 0.1590 | No |
| 42 | Mapk1 | 6978 | 0.084 | 0.1603 | No |
| 43 | Atf1 | 7043 | 0.082 | 0.1610 | No |
| 44 | Map2k6 | 7053 | 0.081 | 0.1646 | No |
| 45 | Mapkap1 | 7227 | 0.076 | 0.1594 | No |
| 46 | Ap2m1 | 7248 | 0.075 | 0.1621 | No |
| 47 | Tsc2 | 7318 | 0.072 | 0.1621 | No |
| 48 | Pdk1 | 7328 | 0.072 | 0.1651 | No |
| 49 | Itpr2 | 7454 | 0.067 | 0.1620 | No |
| 50 | Sfn | 7505 | 0.065 | 0.1627 | No |
| 51 | Cdk2 | 7557 | 0.064 | 0.1632 | No |
| 52 | Plcb1 | 7901 | 0.051 | 0.1480 | No |
| 53 | Mapk9 | 7993 | 0.047 | 0.1456 | No |
| 54 | Cab39l | 8132 | 0.042 | 0.1406 | No |
| 55 | Gsk3b | 8170 | 0.040 | 0.1407 | No |
| 56 | Cdk4 | 8877 | 0.015 | 0.1050 | No |
| 57 | Arhgdia | 8951 | 0.012 | 0.1019 | No |
| 58 | Vav3 | 9023 | 0.009 | 0.0987 | No |
| 59 | Pitx2 | 9483 | -0.007 | 0.0754 | No |
| 60 | Rps6ka1 | 9499 | -0.007 | 0.0749 | No |
| 61 | Sqstm1 | 9579 | -0.010 | 0.0714 | No |
| 62 | Acaca | 10137 | -0.030 | 0.0441 | No |
| 63 | Irak4 | 10360 | -0.039 | 0.0346 | No |
| 64 | Prkag1 | 10411 | -0.041 | 0.0341 | No |
| 65 | Ecsit | 10513 | -0.044 | 0.0310 | No |
| 66 | Cfl1 | 10591 | -0.047 | 0.0294 | No |
| 67 | Mknk2 | 10606 | -0.048 | 0.0310 | No |
| 68 | Ywhab | 11025 | -0.062 | 0.0125 | No |
| 69 | Il2rg | 11838 | -0.094 | -0.0247 | No |
| 70 | Slc2a1 | 11956 | -0.098 | -0.0259 | No |
| 71 | Tnfrsf1a | 12089 | -0.103 | -0.0276 | No |
| 72 | Cxcr4 | 12238 | -0.109 | -0.0298 | No |
| 73 | Tiam1 | 12272 | -0.111 | -0.0261 | No |
| 74 | Them4 | 12350 | -0.114 | -0.0244 | No |
| 75 | Sla | 12363 | -0.115 | -0.0194 | No |
| 76 | Hras | 12554 | -0.123 | -0.0231 | No |
| 77 | Prkar2a | 12595 | -0.124 | -0.0191 | No |
| 78 | Rptor | 12633 | -0.126 | -0.0148 | No |
| 79 | Rit1 | 13527 | -0.162 | -0.0528 | No |
| 80 | Pten | 13685 | -0.169 | -0.0526 | No |
| 81 | Stat2 | 14132 | -0.189 | -0.0662 | No |
| 82 | Ppp1ca | 14444 | -0.203 | -0.0722 | No |
| 83 | Pin1 | 14577 | -0.210 | -0.0687 | No |
| 84 | Cdk1 | 14621 | -0.212 | -0.0605 | No |
| 85 | Mknk1 | 14812 | -0.221 | -0.0594 | No |
| 86 | Akt1s1 | 15381 | -0.249 | -0.0764 | No |
| 87 | Camk4 | 15398 | -0.250 | -0.0650 | No |
| 88 | Pak4 | 15520 | -0.257 | -0.0585 | No |
| 89 | Adcy2 | 15753 | -0.269 | -0.0572 | No |
| 90 | Pla2g12a | 15984 | -0.282 | -0.0552 | No |
| 91 | Dapp1 | 16018 | -0.284 | -0.0429 | No |
| 92 | Pfn1 | 16595 | -0.322 | -0.0568 | No |
| 93 | Csnk2b | 17830 | -0.423 | -0.0995 | No |
| 94 | Dusp3 | 18018 | -0.446 | -0.0871 | No |
| 95 | Prkcb | 18882 | -0.581 | -0.1030 | No |
| 96 | Gna14 | 19425 | -0.845 | -0.0894 | No |
| 97 | Pik3r3 | 19469 | -0.950 | -0.0448 | No |
| 98 | Trib3 | 19471 | -0.954 | 0.0021 | No |