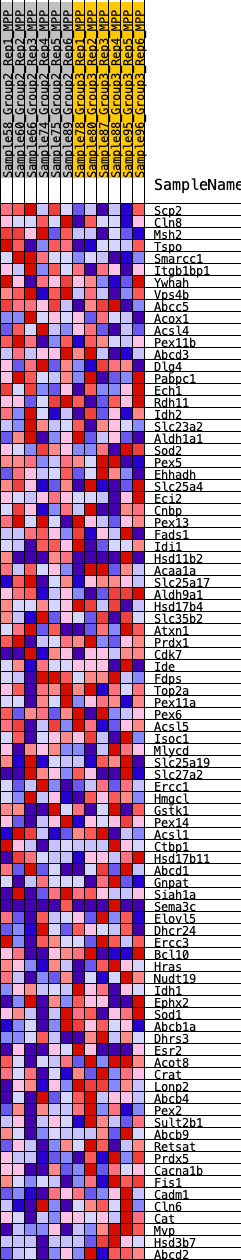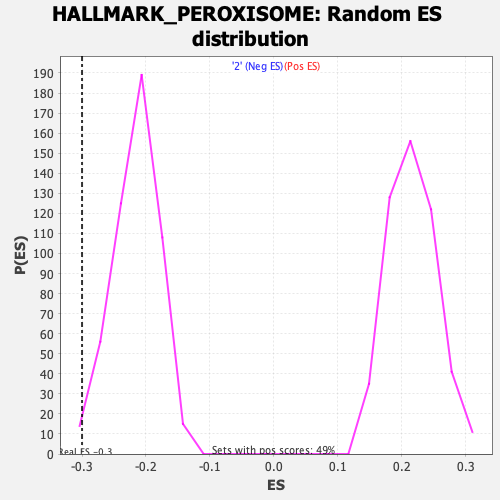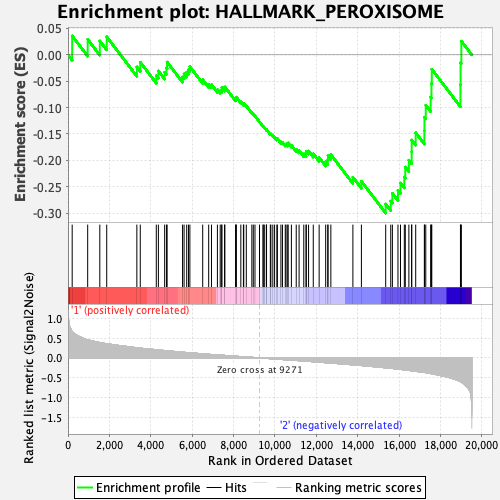
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.29994467 |
| Normalized Enrichment Score (NES) | -1.3881675 |
| Nominal p-value | 0.015779093 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.699 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Scp2 | 202 | 0.663 | 0.0359 | No |
| 2 | Cln8 | 951 | 0.457 | 0.0294 | No |
| 3 | Msh2 | 1537 | 0.388 | 0.0264 | No |
| 4 | Tspo | 1871 | 0.357 | 0.0342 | No |
| 5 | Smarcc1 | 3327 | 0.256 | -0.0228 | No |
| 6 | Itgb1bp1 | 3492 | 0.246 | -0.0140 | No |
| 7 | Ywhah | 4266 | 0.206 | -0.0394 | No |
| 8 | Vps4b | 4367 | 0.201 | -0.0305 | No |
| 9 | Abcc5 | 4670 | 0.186 | -0.0330 | No |
| 10 | Acox1 | 4757 | 0.182 | -0.0247 | No |
| 11 | Acsl4 | 4789 | 0.181 | -0.0136 | No |
| 12 | Pex11b | 5535 | 0.144 | -0.0419 | No |
| 13 | Abcd3 | 5609 | 0.141 | -0.0359 | No |
| 14 | Dlg4 | 5735 | 0.135 | -0.0328 | No |
| 15 | Pabpc1 | 5821 | 0.132 | -0.0280 | No |
| 16 | Ech1 | 5889 | 0.130 | -0.0224 | No |
| 17 | Rdh11 | 6509 | 0.103 | -0.0471 | No |
| 18 | Idh2 | 6798 | 0.091 | -0.0555 | No |
| 19 | Slc23a2 | 6933 | 0.086 | -0.0564 | No |
| 20 | Aldh1a1 | 7219 | 0.076 | -0.0658 | No |
| 21 | Sod2 | 7355 | 0.071 | -0.0678 | No |
| 22 | Pex5 | 7430 | 0.068 | -0.0668 | No |
| 23 | Ehhadh | 7432 | 0.068 | -0.0621 | No |
| 24 | Slc25a4 | 7566 | 0.063 | -0.0645 | No |
| 25 | Eci2 | 7578 | 0.063 | -0.0607 | No |
| 26 | Cnbp | 8097 | 0.043 | -0.0843 | No |
| 27 | Pex13 | 8121 | 0.042 | -0.0825 | No |
| 28 | Fads1 | 8131 | 0.042 | -0.0801 | No |
| 29 | Idi1 | 8351 | 0.033 | -0.0890 | No |
| 30 | Hsd11b2 | 8476 | 0.029 | -0.0934 | No |
| 31 | Acaa1a | 8493 | 0.028 | -0.0922 | No |
| 32 | Slc25a17 | 8615 | 0.023 | -0.0969 | No |
| 33 | Aldh9a1 | 8884 | 0.014 | -0.1097 | No |
| 34 | Hsd17b4 | 8953 | 0.012 | -0.1123 | No |
| 35 | Slc35b2 | 9035 | 0.009 | -0.1159 | No |
| 36 | Atxn1 | 9253 | 0.001 | -0.1270 | No |
| 37 | Prdx1 | 9422 | -0.004 | -0.1353 | No |
| 38 | Cdk7 | 9435 | -0.005 | -0.1356 | No |
| 39 | Ide | 9504 | -0.007 | -0.1386 | No |
| 40 | Fdps | 9592 | -0.011 | -0.1424 | No |
| 41 | Top2a | 9593 | -0.011 | -0.1416 | No |
| 42 | Pex11a | 9776 | -0.017 | -0.1498 | No |
| 43 | Pex6 | 9803 | -0.018 | -0.1499 | No |
| 44 | Acsl5 | 9897 | -0.021 | -0.1533 | No |
| 45 | Isoc1 | 9982 | -0.024 | -0.1559 | No |
| 46 | Mlycd | 10094 | -0.028 | -0.1596 | No |
| 47 | Slc25a19 | 10116 | -0.029 | -0.1587 | No |
| 48 | Slc27a2 | 10288 | -0.036 | -0.1650 | No |
| 49 | Ercc1 | 10363 | -0.039 | -0.1660 | No |
| 50 | Hmgcl | 10498 | -0.044 | -0.1699 | No |
| 51 | Gstk1 | 10537 | -0.045 | -0.1687 | No |
| 52 | Pex14 | 10620 | -0.048 | -0.1695 | No |
| 53 | Acsl1 | 10634 | -0.048 | -0.1668 | No |
| 54 | Ctbp1 | 10794 | -0.054 | -0.1713 | No |
| 55 | Hsd17b11 | 11028 | -0.062 | -0.1789 | No |
| 56 | Abcd1 | 11166 | -0.067 | -0.1813 | No |
| 57 | Gnpat | 11393 | -0.076 | -0.1876 | No |
| 58 | Siah1a | 11501 | -0.080 | -0.1875 | No |
| 59 | Sema3c | 11515 | -0.080 | -0.1826 | No |
| 60 | Elovl5 | 11624 | -0.084 | -0.1822 | No |
| 61 | Dhcr24 | 11851 | -0.094 | -0.1873 | No |
| 62 | Ercc3 | 12135 | -0.105 | -0.1945 | No |
| 63 | Bcl10 | 12451 | -0.118 | -0.2025 | No |
| 64 | Hras | 12554 | -0.123 | -0.1992 | No |
| 65 | Nudt19 | 12563 | -0.123 | -0.1909 | No |
| 66 | Idh1 | 12701 | -0.128 | -0.1890 | No |
| 67 | Ephx2 | 13766 | -0.173 | -0.2317 | No |
| 68 | Sod1 | 14177 | -0.190 | -0.2396 | No |
| 69 | Abcb1a | 15351 | -0.248 | -0.2826 | Yes |
| 70 | Dhrs3 | 15595 | -0.261 | -0.2769 | Yes |
| 71 | Esr2 | 15674 | -0.265 | -0.2624 | Yes |
| 72 | Acot8 | 15944 | -0.280 | -0.2567 | Yes |
| 73 | Crat | 16066 | -0.287 | -0.2428 | Yes |
| 74 | Lonp2 | 16251 | -0.300 | -0.2314 | Yes |
| 75 | Abcb4 | 16298 | -0.303 | -0.2126 | Yes |
| 76 | Pex2 | 16471 | -0.314 | -0.1995 | Yes |
| 77 | Sult2b1 | 16605 | -0.323 | -0.1838 | Yes |
| 78 | Abcb9 | 16608 | -0.323 | -0.1613 | Yes |
| 79 | Retsat | 16801 | -0.337 | -0.1476 | Yes |
| 80 | Prdx5 | 17220 | -0.366 | -0.1436 | Yes |
| 81 | Cacna1b | 17224 | -0.366 | -0.1182 | Yes |
| 82 | Fis1 | 17287 | -0.371 | -0.0954 | Yes |
| 83 | Cadm1 | 17523 | -0.393 | -0.0801 | Yes |
| 84 | Cln6 | 17562 | -0.397 | -0.0543 | Yes |
| 85 | Cat | 17578 | -0.400 | -0.0272 | Yes |
| 86 | Mvp | 18963 | -0.600 | -0.0565 | Yes |
| 87 | Hsd3b7 | 18969 | -0.601 | -0.0147 | Yes |
| 88 | Abcd2 | 19007 | -0.610 | 0.0260 | Yes |