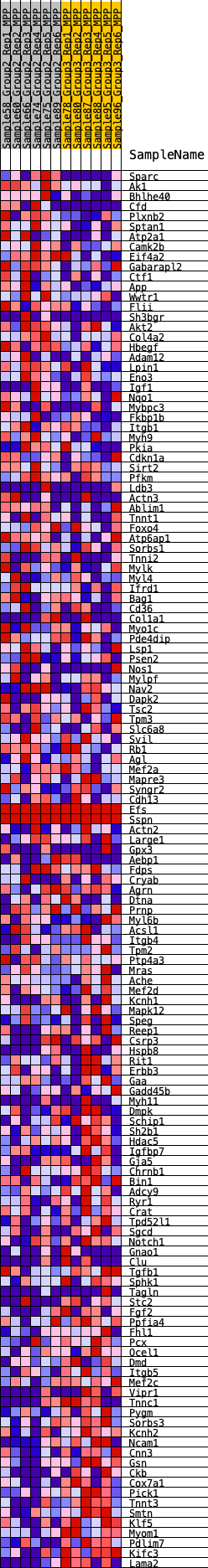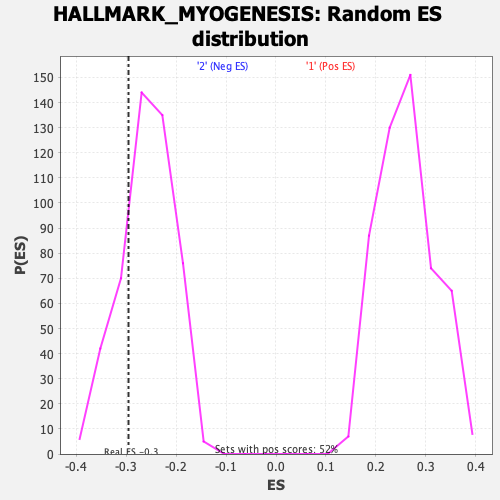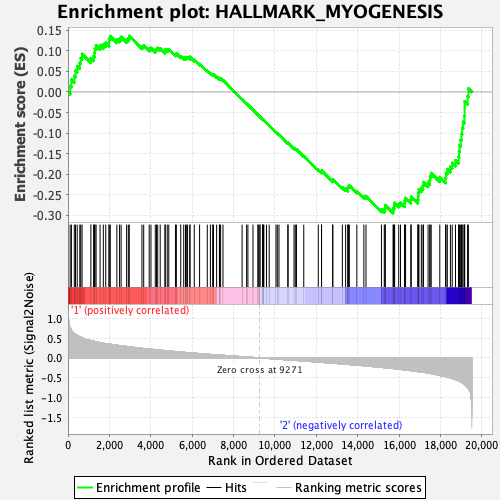
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.29517668 |
| Normalized Enrichment Score (NES) | -1.145016 |
| Nominal p-value | 0.23849373 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.966 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sparc | 104 | 0.766 | 0.0150 | No |
| 2 | Ak1 | 165 | 0.688 | 0.0302 | No |
| 3 | Bhlhe40 | 304 | 0.614 | 0.0394 | No |
| 4 | Cfd | 363 | 0.593 | 0.0521 | No |
| 5 | Plxnb2 | 445 | 0.569 | 0.0631 | No |
| 6 | Sptan1 | 571 | 0.535 | 0.0708 | No |
| 7 | Atp2a1 | 616 | 0.525 | 0.0825 | No |
| 8 | Camk2b | 679 | 0.508 | 0.0928 | No |
| 9 | Eif4a2 | 1105 | 0.438 | 0.0825 | No |
| 10 | Gabarapl2 | 1236 | 0.422 | 0.0870 | No |
| 11 | Ctf1 | 1283 | 0.416 | 0.0957 | No |
| 12 | App | 1302 | 0.414 | 0.1058 | No |
| 13 | Wwtr1 | 1357 | 0.407 | 0.1138 | No |
| 14 | Flii | 1549 | 0.387 | 0.1142 | No |
| 15 | Sh3bgr | 1707 | 0.371 | 0.1160 | No |
| 16 | Akt2 | 1823 | 0.361 | 0.1196 | No |
| 17 | Col4a2 | 1983 | 0.349 | 0.1207 | No |
| 18 | Hbegf | 1987 | 0.349 | 0.1298 | No |
| 19 | Adam12 | 2043 | 0.344 | 0.1361 | No |
| 20 | Lpin1 | 2362 | 0.320 | 0.1282 | No |
| 21 | Eno3 | 2491 | 0.312 | 0.1299 | No |
| 22 | Igf1 | 2566 | 0.307 | 0.1343 | No |
| 23 | Nqo1 | 2833 | 0.289 | 0.1282 | No |
| 24 | Mybpc3 | 2915 | 0.284 | 0.1316 | No |
| 25 | Fkbp1b | 2961 | 0.282 | 0.1368 | No |
| 26 | Itgb1 | 3573 | 0.242 | 0.1116 | No |
| 27 | Myh9 | 3658 | 0.237 | 0.1136 | No |
| 28 | Pkia | 3924 | 0.223 | 0.1059 | No |
| 29 | Cdkn1a | 4008 | 0.219 | 0.1074 | No |
| 30 | Sirt2 | 4217 | 0.209 | 0.1022 | No |
| 31 | Pfkm | 4275 | 0.206 | 0.1047 | No |
| 32 | Ldb3 | 4334 | 0.203 | 0.1071 | No |
| 33 | Actn3 | 4455 | 0.197 | 0.1062 | No |
| 34 | Ablim1 | 4686 | 0.186 | 0.0993 | No |
| 35 | Tnnt1 | 4697 | 0.185 | 0.1037 | No |
| 36 | Foxo4 | 4795 | 0.180 | 0.1034 | No |
| 37 | Atp6ap1 | 4869 | 0.176 | 0.1043 | No |
| 38 | Sorbs1 | 5193 | 0.161 | 0.0919 | No |
| 39 | Tnni2 | 5236 | 0.158 | 0.0940 | No |
| 40 | Mylk | 5448 | 0.148 | 0.0870 | No |
| 41 | Myl4 | 5591 | 0.141 | 0.0835 | No |
| 42 | Ifrd1 | 5678 | 0.138 | 0.0827 | No |
| 43 | Bag1 | 5717 | 0.136 | 0.0843 | No |
| 44 | Cd36 | 5777 | 0.134 | 0.0848 | No |
| 45 | Col1a1 | 5884 | 0.130 | 0.0828 | No |
| 46 | Myo1c | 5896 | 0.129 | 0.0857 | No |
| 47 | Pde4dip | 6101 | 0.121 | 0.0784 | No |
| 48 | Lsp1 | 6358 | 0.109 | 0.0681 | No |
| 49 | Psen2 | 6730 | 0.094 | 0.0514 | No |
| 50 | Nos1 | 6881 | 0.088 | 0.0460 | No |
| 51 | Mylpf | 6991 | 0.083 | 0.0426 | No |
| 52 | Nav2 | 7031 | 0.082 | 0.0428 | No |
| 53 | Dapk2 | 7182 | 0.078 | 0.0371 | No |
| 54 | Tsc2 | 7318 | 0.072 | 0.0320 | No |
| 55 | Tpm3 | 7319 | 0.072 | 0.0340 | No |
| 56 | Slc6a8 | 7374 | 0.070 | 0.0330 | No |
| 57 | Svil | 7489 | 0.066 | 0.0289 | No |
| 58 | Rb1 | 8412 | 0.031 | -0.0178 | No |
| 59 | Agl | 8631 | 0.022 | -0.0285 | No |
| 60 | Mef2a | 8693 | 0.020 | -0.0311 | No |
| 61 | Mapre3 | 8938 | 0.013 | -0.0434 | No |
| 62 | Syngr2 | 9174 | 0.004 | -0.0554 | No |
| 63 | Cdh13 | 9195 | 0.003 | -0.0563 | No |
| 64 | Efs | 9272 | 0.000 | -0.0602 | No |
| 65 | Sspn | 9275 | 0.000 | -0.0604 | No |
| 66 | Actn2 | 9397 | -0.003 | -0.0665 | No |
| 67 | Large1 | 9417 | -0.004 | -0.0674 | No |
| 68 | Gpx3 | 9444 | -0.005 | -0.0686 | No |
| 69 | Aebp1 | 9464 | -0.006 | -0.0694 | No |
| 70 | Fdps | 9592 | -0.011 | -0.0757 | No |
| 71 | Cryab | 9721 | -0.015 | -0.0819 | No |
| 72 | Agrn | 10049 | -0.027 | -0.0981 | No |
| 73 | Dtna | 10113 | -0.029 | -0.1006 | No |
| 74 | Prnp | 10194 | -0.032 | -0.1038 | No |
| 75 | Myl6b | 10622 | -0.048 | -0.1246 | No |
| 76 | Acsl1 | 10634 | -0.048 | -0.1239 | No |
| 77 | Itgb4 | 10921 | -0.059 | -0.1371 | No |
| 78 | Tpm2 | 10983 | -0.060 | -0.1386 | No |
| 79 | Ptp4a3 | 11040 | -0.063 | -0.1398 | No |
| 80 | Mras | 11392 | -0.076 | -0.1559 | No |
| 81 | Ache | 12094 | -0.103 | -0.1894 | No |
| 82 | Mef2d | 12249 | -0.110 | -0.1944 | No |
| 83 | Kcnh1 | 12253 | -0.110 | -0.1916 | No |
| 84 | Mapk12 | 12788 | -0.132 | -0.2157 | No |
| 85 | Speg | 12797 | -0.132 | -0.2126 | No |
| 86 | Reep1 | 13256 | -0.150 | -0.2322 | No |
| 87 | Csrp3 | 13414 | -0.157 | -0.2361 | No |
| 88 | Hspb8 | 13526 | -0.162 | -0.2376 | No |
| 89 | Rit1 | 13527 | -0.162 | -0.2332 | No |
| 90 | Erbb3 | 13547 | -0.164 | -0.2299 | No |
| 91 | Gaa | 13594 | -0.165 | -0.2278 | No |
| 92 | Gadd45b | 13957 | -0.181 | -0.2417 | No |
| 93 | Myh11 | 14296 | -0.196 | -0.2539 | No |
| 94 | Dmpk | 14399 | -0.201 | -0.2539 | No |
| 95 | Schip1 | 15146 | -0.237 | -0.2861 | No |
| 96 | Sh2b1 | 15282 | -0.245 | -0.2865 | No |
| 97 | Hdac5 | 15320 | -0.247 | -0.2819 | No |
| 98 | Igfbp7 | 15328 | -0.247 | -0.2757 | No |
| 99 | Gja5 | 15707 | -0.267 | -0.2881 | Yes |
| 100 | Chrnb1 | 15728 | -0.268 | -0.2820 | Yes |
| 101 | Bin1 | 15762 | -0.270 | -0.2765 | Yes |
| 102 | Adcy9 | 15783 | -0.271 | -0.2704 | Yes |
| 103 | Ryr1 | 15980 | -0.282 | -0.2730 | Yes |
| 104 | Crat | 16066 | -0.287 | -0.2697 | Yes |
| 105 | Tpd52l1 | 16261 | -0.300 | -0.2718 | Yes |
| 106 | Sgcd | 16273 | -0.301 | -0.2643 | Yes |
| 107 | Notch1 | 16312 | -0.304 | -0.2582 | Yes |
| 108 | Gnao1 | 16564 | -0.320 | -0.2627 | Yes |
| 109 | Clu | 16584 | -0.321 | -0.2551 | Yes |
| 110 | Tgfb1 | 16902 | -0.344 | -0.2623 | Yes |
| 111 | Sphk1 | 16923 | -0.346 | -0.2542 | Yes |
| 112 | Tagln | 16929 | -0.346 | -0.2452 | Yes |
| 113 | Stc2 | 16957 | -0.348 | -0.2374 | Yes |
| 114 | Fgf2 | 17073 | -0.355 | -0.2339 | Yes |
| 115 | Ppfia4 | 17153 | -0.361 | -0.2283 | Yes |
| 116 | Fhl1 | 17174 | -0.362 | -0.2197 | Yes |
| 117 | Pcx | 17399 | -0.380 | -0.2212 | Yes |
| 118 | Ocel1 | 17481 | -0.389 | -0.2151 | Yes |
| 119 | Dmd | 17495 | -0.390 | -0.2054 | Yes |
| 120 | Itgb5 | 17549 | -0.395 | -0.1976 | Yes |
| 121 | Mef2c | 17965 | -0.439 | -0.2073 | Yes |
| 122 | Vipr1 | 18244 | -0.473 | -0.2091 | Yes |
| 123 | Tnnc1 | 18264 | -0.476 | -0.1974 | Yes |
| 124 | Pygm | 18328 | -0.483 | -0.1878 | Yes |
| 125 | Sorbs3 | 18477 | -0.504 | -0.1821 | Yes |
| 126 | Kcnh2 | 18565 | -0.518 | -0.1728 | Yes |
| 127 | Ncam1 | 18724 | -0.546 | -0.1664 | Yes |
| 128 | Cnn3 | 18872 | -0.578 | -0.1587 | Yes |
| 129 | Gsn | 18896 | -0.583 | -0.1444 | Yes |
| 130 | Ckb | 18922 | -0.591 | -0.1300 | Yes |
| 131 | Cox7a1 | 18974 | -0.602 | -0.1166 | Yes |
| 132 | Pick1 | 19013 | -0.611 | -0.1023 | Yes |
| 133 | Tnnt3 | 19045 | -0.622 | -0.0874 | Yes |
| 134 | Smtn | 19090 | -0.637 | -0.0727 | Yes |
| 135 | Klf5 | 19153 | -0.670 | -0.0581 | Yes |
| 136 | Myom1 | 19164 | -0.674 | -0.0407 | Yes |
| 137 | Pdlim7 | 19165 | -0.674 | -0.0228 | Yes |
| 138 | Kifc3 | 19309 | -0.754 | -0.0101 | Yes |
| 139 | Lama2 | 19347 | -0.774 | 0.0085 | Yes |