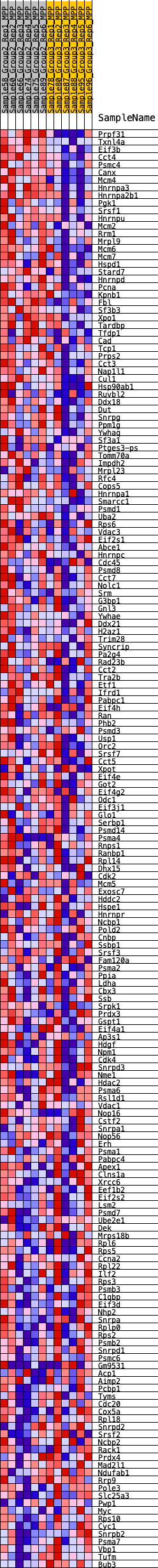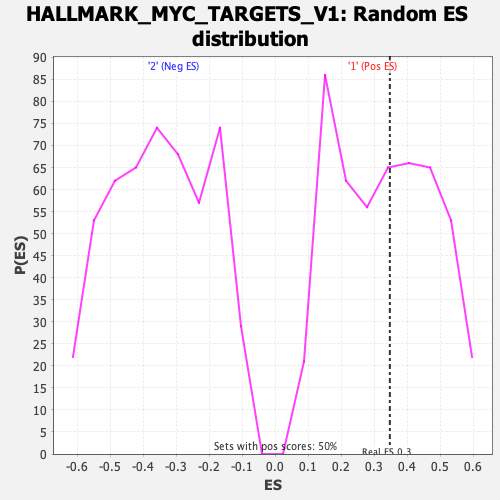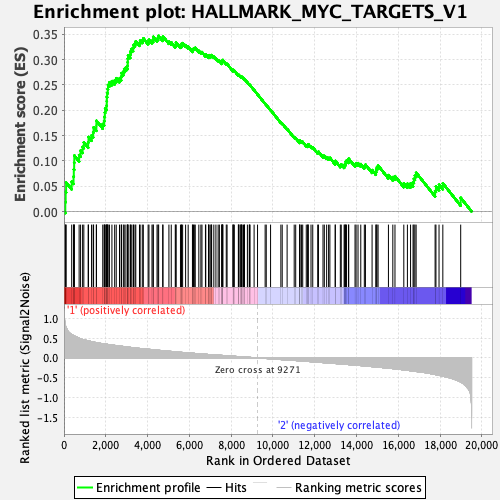
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_MYC_TARGETS_V1 |
| Enrichment Score (ES) | 0.34685624 |
| Normalized Enrichment Score (NES) | 1.044293 |
| Nominal p-value | 0.4657258 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prpf31 | 63 | 0.851 | 0.0187 | Yes |
| 2 | Txnl4a | 80 | 0.800 | 0.0386 | Yes |
| 3 | Eif3b | 102 | 0.767 | 0.0574 | Yes |
| 4 | Cct4 | 369 | 0.591 | 0.0589 | Yes |
| 5 | Psmc4 | 457 | 0.565 | 0.0690 | Yes |
| 6 | Canx | 469 | 0.563 | 0.0830 | Yes |
| 7 | Mcm4 | 487 | 0.558 | 0.0965 | Yes |
| 8 | Hnrnpa3 | 494 | 0.555 | 0.1106 | Yes |
| 9 | Hnrnpa2b1 | 724 | 0.497 | 0.1116 | Yes |
| 10 | Pgk1 | 794 | 0.481 | 0.1204 | Yes |
| 11 | Srsf1 | 891 | 0.466 | 0.1275 | Yes |
| 12 | Hnrnpu | 949 | 0.457 | 0.1364 | Yes |
| 13 | Mcm2 | 1156 | 0.431 | 0.1369 | Yes |
| 14 | Rrm1 | 1173 | 0.429 | 0.1472 | Yes |
| 15 | Mrpl9 | 1319 | 0.412 | 0.1503 | Yes |
| 16 | Mcm6 | 1397 | 0.402 | 0.1567 | Yes |
| 17 | Mcm7 | 1416 | 0.399 | 0.1661 | Yes |
| 18 | Hspd1 | 1550 | 0.387 | 0.1692 | Yes |
| 19 | Stard7 | 1551 | 0.387 | 0.1793 | Yes |
| 20 | Hnrnpd | 1855 | 0.358 | 0.1728 | Yes |
| 21 | Pcna | 1921 | 0.355 | 0.1786 | Yes |
| 22 | Kpnb1 | 1936 | 0.353 | 0.1871 | Yes |
| 23 | Fbl | 1956 | 0.352 | 0.1952 | Yes |
| 24 | Sf3b3 | 1970 | 0.351 | 0.2036 | Yes |
| 25 | Xpo1 | 2039 | 0.345 | 0.2090 | Yes |
| 26 | Tardbp | 2047 | 0.344 | 0.2175 | Yes |
| 27 | Tfdp1 | 2051 | 0.344 | 0.2263 | Yes |
| 28 | Cad | 2066 | 0.343 | 0.2344 | Yes |
| 29 | Tcp1 | 2094 | 0.341 | 0.2418 | Yes |
| 30 | Prps2 | 2107 | 0.340 | 0.2500 | Yes |
| 31 | Cct3 | 2175 | 0.335 | 0.2552 | Yes |
| 32 | Nap1l1 | 2294 | 0.325 | 0.2575 | Yes |
| 33 | Cul1 | 2426 | 0.316 | 0.2589 | Yes |
| 34 | Hsp90ab1 | 2500 | 0.311 | 0.2632 | Yes |
| 35 | Ruvbl2 | 2657 | 0.301 | 0.2629 | Yes |
| 36 | Ddx18 | 2737 | 0.296 | 0.2665 | Yes |
| 37 | Dut | 2753 | 0.295 | 0.2733 | Yes |
| 38 | Snrpg | 2840 | 0.288 | 0.2763 | Yes |
| 39 | Ppm1g | 2883 | 0.286 | 0.2816 | Yes |
| 40 | Ywhaq | 2964 | 0.281 | 0.2847 | Yes |
| 41 | Sf3a1 | 3040 | 0.277 | 0.2880 | Yes |
| 42 | Ptges3-ps | 3048 | 0.276 | 0.2947 | Yes |
| 43 | Tomm70a | 3054 | 0.275 | 0.3016 | Yes |
| 44 | Impdh2 | 3066 | 0.275 | 0.3081 | Yes |
| 45 | Mrpl23 | 3169 | 0.267 | 0.3098 | Yes |
| 46 | Rfc4 | 3181 | 0.266 | 0.3161 | Yes |
| 47 | Cops5 | 3220 | 0.264 | 0.3209 | Yes |
| 48 | Hnrnpa1 | 3307 | 0.257 | 0.3231 | Yes |
| 49 | Smarcc1 | 3327 | 0.256 | 0.3288 | Yes |
| 50 | Psmd1 | 3397 | 0.251 | 0.3317 | Yes |
| 51 | Uba2 | 3436 | 0.249 | 0.3362 | Yes |
| 52 | Rps6 | 3613 | 0.240 | 0.3333 | Yes |
| 53 | Vdac3 | 3645 | 0.238 | 0.3378 | Yes |
| 54 | Eif2s1 | 3747 | 0.232 | 0.3386 | Yes |
| 55 | Abce1 | 3797 | 0.230 | 0.3420 | Yes |
| 56 | Hnrnpc | 4026 | 0.218 | 0.3359 | Yes |
| 57 | Cdc45 | 4067 | 0.216 | 0.3394 | Yes |
| 58 | Psmd8 | 4212 | 0.209 | 0.3373 | Yes |
| 59 | Cct7 | 4268 | 0.206 | 0.3398 | Yes |
| 60 | Nolc1 | 4271 | 0.206 | 0.3451 | Yes |
| 61 | Srm | 4454 | 0.197 | 0.3407 | Yes |
| 62 | G3bp1 | 4502 | 0.194 | 0.3433 | Yes |
| 63 | Gnl3 | 4531 | 0.192 | 0.3469 | Yes |
| 64 | Ywhae | 4725 | 0.184 | 0.3416 | No |
| 65 | Ddx21 | 4738 | 0.183 | 0.3457 | No |
| 66 | H2az1 | 5022 | 0.169 | 0.3355 | No |
| 67 | Trim28 | 5134 | 0.164 | 0.3340 | No |
| 68 | Syncrip | 5326 | 0.154 | 0.3281 | No |
| 69 | Pa2g4 | 5356 | 0.152 | 0.3305 | No |
| 70 | Rad23b | 5362 | 0.152 | 0.3342 | No |
| 71 | Cct2 | 5575 | 0.142 | 0.3269 | No |
| 72 | Tra2b | 5626 | 0.140 | 0.3279 | No |
| 73 | Etf1 | 5628 | 0.140 | 0.3315 | No |
| 74 | Ifrd1 | 5678 | 0.138 | 0.3325 | No |
| 75 | Pabpc1 | 5821 | 0.132 | 0.3285 | No |
| 76 | Eif4h | 5944 | 0.127 | 0.3255 | No |
| 77 | Ran | 6148 | 0.119 | 0.3181 | No |
| 78 | Phb2 | 6162 | 0.118 | 0.3205 | No |
| 79 | Psmd3 | 6200 | 0.117 | 0.3216 | No |
| 80 | Usp1 | 6238 | 0.115 | 0.3226 | No |
| 81 | Orc2 | 6282 | 0.113 | 0.3233 | No |
| 82 | Srsf7 | 6454 | 0.105 | 0.3172 | No |
| 83 | Cct5 | 6552 | 0.101 | 0.3148 | No |
| 84 | Xpot | 6604 | 0.099 | 0.3147 | No |
| 85 | Eif4e | 6771 | 0.092 | 0.3085 | No |
| 86 | Got2 | 6786 | 0.091 | 0.3102 | No |
| 87 | Eif4g2 | 6918 | 0.086 | 0.3056 | No |
| 88 | Odc1 | 6922 | 0.086 | 0.3077 | No |
| 89 | Eif3j1 | 6947 | 0.086 | 0.3087 | No |
| 90 | Glo1 | 7029 | 0.082 | 0.3066 | No |
| 91 | Serbp1 | 7049 | 0.081 | 0.3077 | No |
| 92 | Psmd14 | 7067 | 0.081 | 0.3089 | No |
| 93 | Psma4 | 7172 | 0.078 | 0.3056 | No |
| 94 | Rnps1 | 7272 | 0.074 | 0.3023 | No |
| 95 | Ranbp1 | 7378 | 0.070 | 0.2987 | No |
| 96 | Rpl14 | 7437 | 0.068 | 0.2975 | No |
| 97 | Dhx15 | 7556 | 0.064 | 0.2930 | No |
| 98 | Cdk2 | 7557 | 0.064 | 0.2947 | No |
| 99 | Mcm5 | 7563 | 0.064 | 0.2961 | No |
| 100 | Exosc7 | 7592 | 0.062 | 0.2962 | No |
| 101 | Hddc2 | 7597 | 0.062 | 0.2976 | No |
| 102 | Hspe1 | 7598 | 0.062 | 0.2992 | No |
| 103 | Hnrnpr | 7767 | 0.056 | 0.2920 | No |
| 104 | Ncbp1 | 7806 | 0.054 | 0.2914 | No |
| 105 | Pold2 | 8091 | 0.044 | 0.2779 | No |
| 106 | Cnbp | 8097 | 0.043 | 0.2787 | No |
| 107 | Ssbp1 | 8101 | 0.043 | 0.2797 | No |
| 108 | Srsf3 | 8153 | 0.041 | 0.2781 | No |
| 109 | Fam120a | 8346 | 0.033 | 0.2691 | No |
| 110 | Psma2 | 8360 | 0.033 | 0.2692 | No |
| 111 | Ppia | 8368 | 0.033 | 0.2697 | No |
| 112 | Ldha | 8434 | 0.030 | 0.2671 | No |
| 113 | Cbx3 | 8473 | 0.029 | 0.2659 | No |
| 114 | Ssb | 8483 | 0.029 | 0.2662 | No |
| 115 | Srpk1 | 8494 | 0.028 | 0.2664 | No |
| 116 | Prdx3 | 8528 | 0.026 | 0.2654 | No |
| 117 | Gspt1 | 8592 | 0.024 | 0.2627 | No |
| 118 | Eif4a1 | 8636 | 0.022 | 0.2611 | No |
| 119 | Ap3s1 | 8638 | 0.022 | 0.2616 | No |
| 120 | Hdgf | 8779 | 0.017 | 0.2548 | No |
| 121 | Npm1 | 8869 | 0.015 | 0.2506 | No |
| 122 | Cdk4 | 8877 | 0.015 | 0.2506 | No |
| 123 | Snrpd3 | 8906 | 0.014 | 0.2495 | No |
| 124 | Nme1 | 9100 | 0.007 | 0.2397 | No |
| 125 | Hdac2 | 9264 | 0.000 | 0.2313 | No |
| 126 | Psma6 | 9628 | -0.012 | 0.2128 | No |
| 127 | Rsl1d1 | 9681 | -0.013 | 0.2105 | No |
| 128 | Vdac1 | 9888 | -0.021 | 0.2003 | No |
| 129 | Nop16 | 10381 | -0.040 | 0.1759 | No |
| 130 | Cstf2 | 10445 | -0.042 | 0.1737 | No |
| 131 | Snrpa1 | 10680 | -0.050 | 0.1629 | No |
| 132 | Nop56 | 11019 | -0.062 | 0.1470 | No |
| 133 | Erh | 11091 | -0.064 | 0.1450 | No |
| 134 | Psma1 | 11280 | -0.072 | 0.1371 | No |
| 135 | Pabpc4 | 11284 | -0.072 | 0.1388 | No |
| 136 | Apex1 | 11293 | -0.072 | 0.1403 | No |
| 137 | Clns1a | 11372 | -0.075 | 0.1382 | No |
| 138 | Xrcc6 | 11427 | -0.077 | 0.1374 | No |
| 139 | Eef1b2 | 11597 | -0.083 | 0.1308 | No |
| 140 | Eif2s2 | 11661 | -0.086 | 0.1298 | No |
| 141 | Lsm2 | 11680 | -0.087 | 0.1311 | No |
| 142 | Psmd7 | 11692 | -0.088 | 0.1328 | No |
| 143 | Ube2e1 | 11824 | -0.094 | 0.1284 | No |
| 144 | Dek | 11907 | -0.096 | 0.1267 | No |
| 145 | Mrps18b | 12145 | -0.105 | 0.1171 | No |
| 146 | Rpl6 | 12180 | -0.107 | 0.1181 | No |
| 147 | Rps5 | 12398 | -0.117 | 0.1099 | No |
| 148 | Ccna2 | 12461 | -0.119 | 0.1098 | No |
| 149 | Rpl22 | 12572 | -0.124 | 0.1073 | No |
| 150 | Ilf2 | 12666 | -0.127 | 0.1058 | No |
| 151 | Rps3 | 12727 | -0.130 | 0.1060 | No |
| 152 | Psmb3 | 12983 | -0.139 | 0.0964 | No |
| 153 | C1qbp | 12986 | -0.140 | 0.0999 | No |
| 154 | Eif3d | 13227 | -0.149 | 0.0914 | No |
| 155 | Nhp2 | 13271 | -0.151 | 0.0930 | No |
| 156 | Snrpa | 13409 | -0.157 | 0.0900 | No |
| 157 | Rplp0 | 13435 | -0.158 | 0.0928 | No |
| 158 | Rps2 | 13477 | -0.160 | 0.0948 | No |
| 159 | Psmb2 | 13485 | -0.160 | 0.0986 | No |
| 160 | Snrpd1 | 13525 | -0.162 | 0.1008 | No |
| 161 | Psmc6 | 13622 | -0.167 | 0.1001 | No |
| 162 | Gm9531 | 13627 | -0.167 | 0.1042 | No |
| 163 | Acp1 | 13931 | -0.180 | 0.0932 | No |
| 164 | Aimp2 | 13981 | -0.182 | 0.0954 | No |
| 165 | Pcbp1 | 14074 | -0.186 | 0.0954 | No |
| 166 | Tyms | 14206 | -0.192 | 0.0936 | No |
| 167 | Cdc20 | 14376 | -0.199 | 0.0900 | No |
| 168 | Cox5a | 14429 | -0.203 | 0.0926 | No |
| 169 | Rpl18 | 14747 | -0.218 | 0.0818 | No |
| 170 | Snrpd2 | 14928 | -0.226 | 0.0783 | No |
| 171 | Srsf2 | 14946 | -0.227 | 0.0833 | No |
| 172 | Ncbp2 | 14980 | -0.229 | 0.0875 | No |
| 173 | Rack1 | 15034 | -0.232 | 0.0908 | No |
| 174 | Prdx4 | 15530 | -0.258 | 0.0718 | No |
| 175 | Mad2l1 | 15740 | -0.269 | 0.0680 | No |
| 176 | Ndufab1 | 15843 | -0.274 | 0.0698 | No |
| 177 | Rrp9 | 16266 | -0.301 | 0.0557 | No |
| 178 | Pole3 | 16438 | -0.312 | 0.0549 | No |
| 179 | Slc25a3 | 16589 | -0.321 | 0.0555 | No |
| 180 | Pwp1 | 16709 | -0.331 | 0.0579 | No |
| 181 | Myc | 16749 | -0.334 | 0.0645 | No |
| 182 | Rps10 | 16798 | -0.337 | 0.0707 | No |
| 183 | Cyc1 | 16855 | -0.340 | 0.0767 | No |
| 184 | Snrpb2 | 17764 | -0.417 | 0.0405 | No |
| 185 | Psma7 | 17804 | -0.421 | 0.0493 | No |
| 186 | Vbp1 | 17952 | -0.438 | 0.0531 | No |
| 187 | Tufm | 18134 | -0.460 | 0.0556 | No |
| 188 | Bub3 | 18989 | -0.605 | 0.0271 | No |