Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
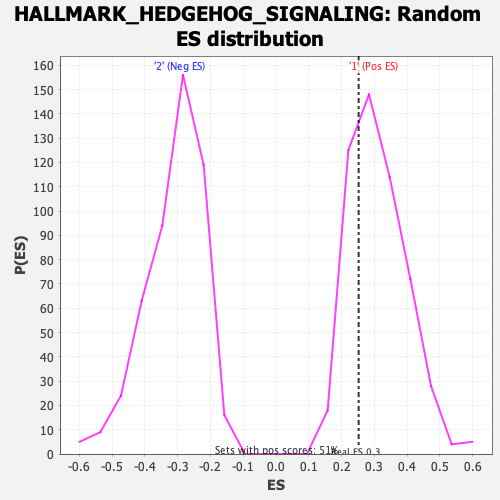
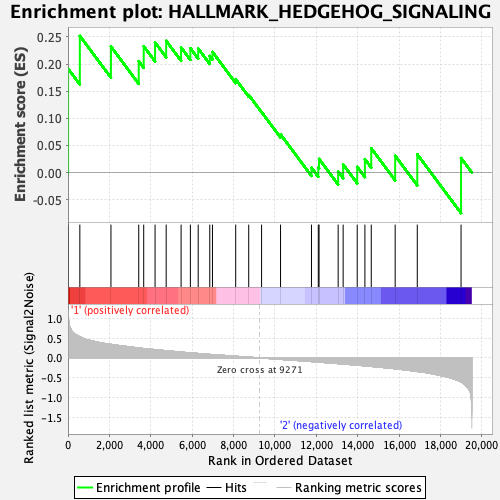
| Enrichment Score (ES) | 0.25204682 |
| Normalized Enrichment Score (NES) | 0.80489856 |
| Nominal p-value | 0.7217899 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
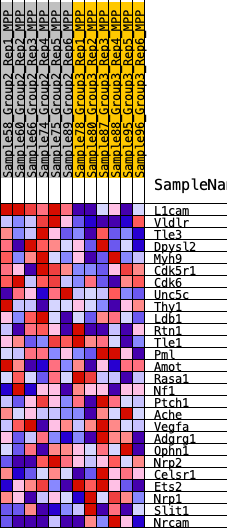
| 1 | L1cam | 4 | 1.144 | 0.1916 | Yes |
| 2 | Vldlr | 572 | 0.535 | 0.2520 | Yes |
| 3 | Tle3 | 2075 | 0.343 | 0.2324 | No |
| 4 | Dpysl2 | 3418 | 0.250 | 0.2054 | No |
| 5 | Myh9 | 3658 | 0.237 | 0.2329 | No |
| 6 | Cdk5r1 | 4207 | 0.210 | 0.2399 | No |
| 7 | Cdk6 | 4744 | 0.183 | 0.2430 | No |
| 8 | Unc5c | 5465 | 0.147 | 0.2307 | No |
| 9 | Thy1 | 5913 | 0.128 | 0.2293 | No |
| 10 | Ldb1 | 6291 | 0.112 | 0.2288 | No |
| 11 | Rtn1 | 6850 | 0.089 | 0.2150 | No |
| 12 | Tle1 | 6980 | 0.084 | 0.2225 | No |
| 13 | Pml | 8103 | 0.043 | 0.1721 | No |
| 14 | Amot | 8732 | 0.019 | 0.1430 | No |
| 15 | Rasa1 | 9354 | -0.002 | 0.1115 | No |
| 16 | Nf1 | 10270 | -0.035 | 0.0704 | No |
| 17 | Ptch1 | 11764 | -0.091 | 0.0090 | No |
| 18 | Ache | 12094 | -0.103 | 0.0095 | No |
| 19 | Vegfa | 12131 | -0.105 | 0.0252 | No |
| 20 | Adgrg1 | 13054 | -0.143 | 0.0018 | No |
| 21 | Ophn1 | 13295 | -0.152 | 0.0150 | No |
| 22 | Nrp2 | 13975 | -0.182 | 0.0105 | No |
| 23 | Celsr1 | 14346 | -0.198 | 0.0247 | No |
| 24 | Ets2 | 14653 | -0.213 | 0.0448 | No |
| 25 | Nrp1 | 15809 | -0.272 | 0.0310 | No |
| 26 | Slit1 | 16877 | -0.342 | 0.0336 | No |
| 27 | Nrcam | 18992 | -0.606 | 0.0267 | No |