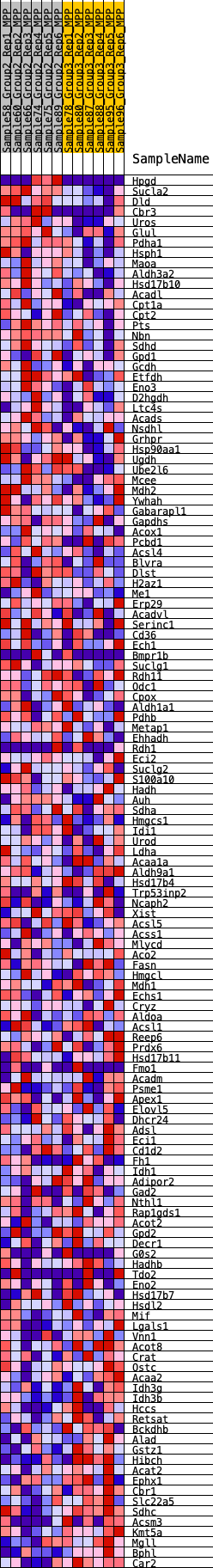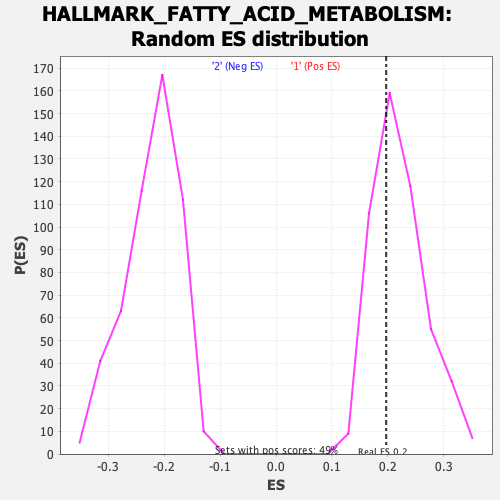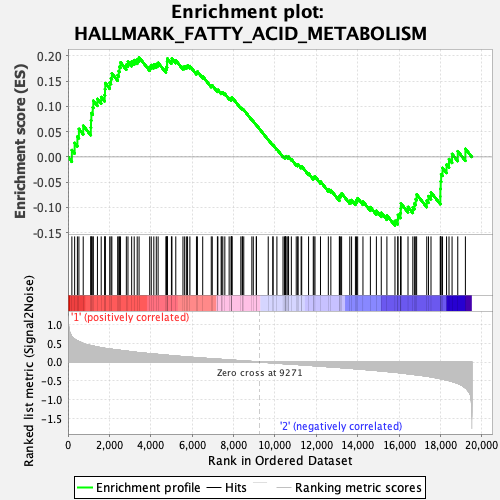
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | 0.19698669 |
| Normalized Enrichment Score (NES) | 0.8920712 |
| Nominal p-value | 0.6255144 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hpgd | 187 | 0.674 | 0.0136 | Yes |
| 2 | Sucla2 | 321 | 0.609 | 0.0277 | Yes |
| 3 | Dld | 454 | 0.567 | 0.0404 | Yes |
| 4 | Cbr3 | 527 | 0.545 | 0.0555 | Yes |
| 5 | Uros | 735 | 0.495 | 0.0619 | Yes |
| 6 | Glul | 1103 | 0.438 | 0.0580 | Yes |
| 7 | Pdha1 | 1112 | 0.437 | 0.0726 | Yes |
| 8 | Hsph1 | 1128 | 0.435 | 0.0869 | Yes |
| 9 | Maoa | 1196 | 0.426 | 0.0981 | Yes |
| 10 | Aldh3a2 | 1222 | 0.423 | 0.1114 | Yes |
| 11 | Hsd17b10 | 1422 | 0.398 | 0.1148 | Yes |
| 12 | Acadl | 1604 | 0.381 | 0.1186 | Yes |
| 13 | Cpt1a | 1773 | 0.365 | 0.1225 | Yes |
| 14 | Cpt2 | 1788 | 0.363 | 0.1343 | Yes |
| 15 | Pts | 1801 | 0.362 | 0.1462 | Yes |
| 16 | Nbn | 2016 | 0.347 | 0.1471 | Yes |
| 17 | Sdhd | 2077 | 0.343 | 0.1558 | Yes |
| 18 | Gpd1 | 2119 | 0.339 | 0.1653 | Yes |
| 19 | Gcdh | 2401 | 0.318 | 0.1618 | Yes |
| 20 | Etfdh | 2454 | 0.315 | 0.1700 | Yes |
| 21 | Eno3 | 2491 | 0.312 | 0.1789 | Yes |
| 22 | D2hgdh | 2538 | 0.310 | 0.1871 | Yes |
| 23 | Ltc4s | 2819 | 0.291 | 0.1827 | Yes |
| 24 | Acads | 2897 | 0.285 | 0.1886 | Yes |
| 25 | Nsdhl | 3076 | 0.274 | 0.1888 | Yes |
| 26 | Grhpr | 3199 | 0.265 | 0.1917 | Yes |
| 27 | Hsp90aa1 | 3344 | 0.255 | 0.1930 | Yes |
| 28 | Ugdh | 3434 | 0.249 | 0.1970 | Yes |
| 29 | Ube2l6 | 3948 | 0.222 | 0.1781 | No |
| 30 | Mcee | 4023 | 0.219 | 0.1819 | No |
| 31 | Mdh2 | 4147 | 0.213 | 0.1828 | No |
| 32 | Ywhah | 4266 | 0.206 | 0.1839 | No |
| 33 | Gabarapl1 | 4354 | 0.202 | 0.1863 | No |
| 34 | Gapdhs | 4731 | 0.183 | 0.1732 | No |
| 35 | Acox1 | 4757 | 0.182 | 0.1782 | No |
| 36 | Pcbd1 | 4788 | 0.181 | 0.1829 | No |
| 37 | Acsl4 | 4789 | 0.181 | 0.1891 | No |
| 38 | Blvra | 4801 | 0.180 | 0.1948 | No |
| 39 | Dlst | 4998 | 0.171 | 0.1905 | No |
| 40 | H2az1 | 5022 | 0.169 | 0.1952 | No |
| 41 | Me1 | 5208 | 0.160 | 0.1911 | No |
| 42 | Erp29 | 5555 | 0.143 | 0.1782 | No |
| 43 | Acadvl | 5637 | 0.140 | 0.1788 | No |
| 44 | Serinc1 | 5719 | 0.136 | 0.1793 | No |
| 45 | Cd36 | 5777 | 0.134 | 0.1810 | No |
| 46 | Ech1 | 5889 | 0.130 | 0.1797 | No |
| 47 | Bmpr1b | 6207 | 0.116 | 0.1674 | No |
| 48 | Suclg1 | 6252 | 0.114 | 0.1690 | No |
| 49 | Rdh11 | 6509 | 0.103 | 0.1594 | No |
| 50 | Odc1 | 6922 | 0.086 | 0.1411 | No |
| 51 | Cpox | 6970 | 0.084 | 0.1416 | No |
| 52 | Aldh1a1 | 7219 | 0.076 | 0.1314 | No |
| 53 | Pdhb | 7242 | 0.075 | 0.1328 | No |
| 54 | Metap1 | 7400 | 0.069 | 0.1271 | No |
| 55 | Ehhadh | 7432 | 0.068 | 0.1279 | No |
| 56 | Rdh1 | 7481 | 0.066 | 0.1277 | No |
| 57 | Eci2 | 7578 | 0.063 | 0.1249 | No |
| 58 | Suclg2 | 7786 | 0.055 | 0.1161 | No |
| 59 | S100a10 | 7878 | 0.052 | 0.1132 | No |
| 60 | Hadh | 7898 | 0.051 | 0.1140 | No |
| 61 | Auh | 7912 | 0.050 | 0.1150 | No |
| 62 | Sdha | 7915 | 0.050 | 0.1167 | No |
| 63 | Hmgcs1 | 7921 | 0.050 | 0.1181 | No |
| 64 | Idi1 | 8351 | 0.033 | 0.0971 | No |
| 65 | Urod | 8415 | 0.031 | 0.0950 | No |
| 66 | Ldha | 8434 | 0.030 | 0.0951 | No |
| 67 | Acaa1a | 8493 | 0.028 | 0.0930 | No |
| 68 | Aldh9a1 | 8884 | 0.014 | 0.0734 | No |
| 69 | Hsd17b4 | 8953 | 0.012 | 0.0703 | No |
| 70 | Trp53inp2 | 9097 | 0.007 | 0.0632 | No |
| 71 | Ncaph2 | 9106 | 0.007 | 0.0630 | No |
| 72 | Xist | 9675 | -0.013 | 0.0341 | No |
| 73 | Acsl5 | 9897 | -0.021 | 0.0234 | No |
| 74 | Acss1 | 9910 | -0.021 | 0.0236 | No |
| 75 | Mlycd | 10094 | -0.028 | 0.0151 | No |
| 76 | Aco2 | 10390 | -0.040 | 0.0012 | No |
| 77 | Fasn | 10459 | -0.042 | -0.0008 | No |
| 78 | Hmgcl | 10498 | -0.044 | -0.0013 | No |
| 79 | Mdh1 | 10503 | -0.044 | 0.0000 | No |
| 80 | Echs1 | 10515 | -0.044 | 0.0010 | No |
| 81 | Cryz | 10538 | -0.045 | 0.0014 | No |
| 82 | Aldoa | 10628 | -0.048 | -0.0015 | No |
| 83 | Acsl1 | 10634 | -0.048 | -0.0001 | No |
| 84 | Reep6 | 10659 | -0.049 | 0.0003 | No |
| 85 | Prdx6 | 10792 | -0.054 | -0.0046 | No |
| 86 | Hsd17b11 | 11028 | -0.062 | -0.0146 | No |
| 87 | Fmo1 | 11099 | -0.065 | -0.0160 | No |
| 88 | Acadm | 11117 | -0.065 | -0.0146 | No |
| 89 | Psme1 | 11265 | -0.071 | -0.0197 | No |
| 90 | Apex1 | 11293 | -0.072 | -0.0186 | No |
| 91 | Elovl5 | 11624 | -0.084 | -0.0328 | No |
| 92 | Dhcr24 | 11851 | -0.094 | -0.0412 | No |
| 93 | Adsl | 11884 | -0.095 | -0.0396 | No |
| 94 | Eci1 | 11934 | -0.097 | -0.0387 | No |
| 95 | Cd1d2 | 12200 | -0.108 | -0.0487 | No |
| 96 | Fh1 | 12579 | -0.124 | -0.0639 | No |
| 97 | Idh1 | 12701 | -0.128 | -0.0658 | No |
| 98 | Adipor2 | 13111 | -0.145 | -0.0819 | No |
| 99 | Gad2 | 13119 | -0.145 | -0.0772 | No |
| 100 | Nthl1 | 13174 | -0.148 | -0.0749 | No |
| 101 | Rap1gds1 | 13221 | -0.148 | -0.0722 | No |
| 102 | Acot2 | 13616 | -0.167 | -0.0868 | No |
| 103 | Gpd2 | 13703 | -0.170 | -0.0854 | No |
| 104 | Decr1 | 13888 | -0.178 | -0.0887 | No |
| 105 | G0s2 | 13942 | -0.181 | -0.0852 | No |
| 106 | Hadhb | 13996 | -0.183 | -0.0817 | No |
| 107 | Tdo2 | 14252 | -0.194 | -0.0882 | No |
| 108 | Eno2 | 14611 | -0.211 | -0.0994 | No |
| 109 | Hsd17b7 | 14901 | -0.225 | -0.1065 | No |
| 110 | Hsdl2 | 15137 | -0.237 | -0.1105 | No |
| 111 | Mif | 15409 | -0.251 | -0.1159 | No |
| 112 | Lgals1 | 15799 | -0.271 | -0.1266 | No |
| 113 | Vnn1 | 15935 | -0.280 | -0.1239 | No |
| 114 | Acot8 | 15944 | -0.280 | -0.1147 | No |
| 115 | Crat | 16066 | -0.287 | -0.1111 | No |
| 116 | Ostc | 16080 | -0.288 | -0.1018 | No |
| 117 | Acaa2 | 16089 | -0.289 | -0.0922 | No |
| 118 | Idh3g | 16429 | -0.311 | -0.0990 | No |
| 119 | Idh3b | 16662 | -0.327 | -0.0997 | No |
| 120 | Hccs | 16738 | -0.333 | -0.0921 | No |
| 121 | Retsat | 16801 | -0.337 | -0.0837 | No |
| 122 | Bckdhb | 16846 | -0.340 | -0.0743 | No |
| 123 | Alad | 17335 | -0.375 | -0.0866 | No |
| 124 | Gstz1 | 17418 | -0.382 | -0.0776 | No |
| 125 | Hibch | 17540 | -0.394 | -0.0703 | No |
| 126 | Acat2 | 17991 | -0.443 | -0.0782 | No |
| 127 | Ephx1 | 17993 | -0.443 | -0.0630 | No |
| 128 | Cbr1 | 18009 | -0.445 | -0.0485 | No |
| 129 | Slc22a5 | 18029 | -0.448 | -0.0340 | No |
| 130 | Sdhc | 18097 | -0.456 | -0.0218 | No |
| 131 | Acsm3 | 18292 | -0.479 | -0.0153 | No |
| 132 | Kmt5a | 18410 | -0.494 | -0.0043 | No |
| 133 | Mgll | 18558 | -0.516 | 0.0059 | No |
| 134 | Bphl | 18833 | -0.568 | 0.0113 | No |
| 135 | Car2 | 19202 | -0.687 | 0.0160 | No |