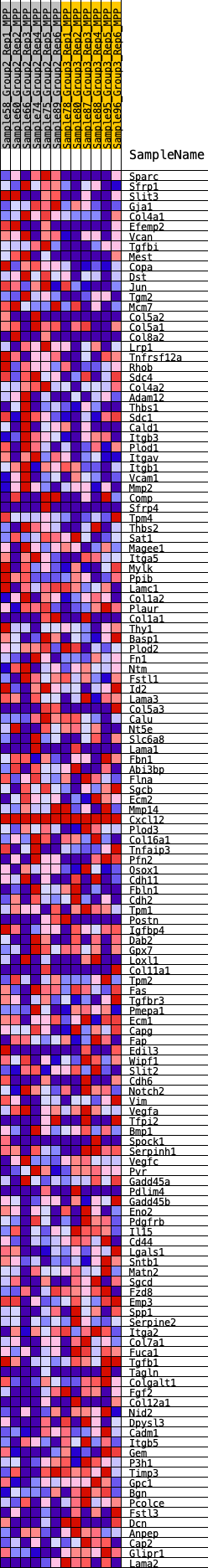
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
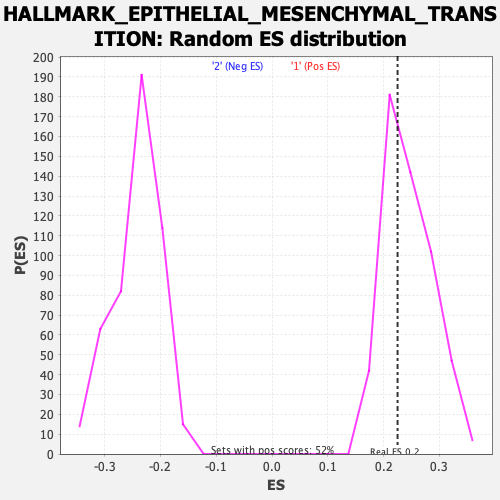
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.22504953 |
| Normalized Enrichment Score (NES) | 0.92225546 |
| Nominal p-value | 0.61804223 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sparc | 104 | 0.766 | 0.0172 | Yes |
| 2 | Sfrp1 | 170 | 0.685 | 0.0341 | Yes |
| 3 | Slit3 | 213 | 0.655 | 0.0512 | Yes |
| 4 | Gja1 | 305 | 0.614 | 0.0647 | Yes |
| 5 | Col4a1 | 396 | 0.581 | 0.0771 | Yes |
| 6 | Efemp2 | 403 | 0.579 | 0.0939 | Yes |
| 7 | Vcan | 466 | 0.563 | 0.1073 | Yes |
| 8 | Tgfbi | 475 | 0.561 | 0.1234 | Yes |
| 9 | Mest | 684 | 0.506 | 0.1276 | Yes |
| 10 | Copa | 696 | 0.504 | 0.1419 | Yes |
| 11 | Dst | 714 | 0.499 | 0.1558 | Yes |
| 12 | Jun | 923 | 0.462 | 0.1587 | Yes |
| 13 | Tgm2 | 1071 | 0.442 | 0.1642 | Yes |
| 14 | Mcm7 | 1416 | 0.399 | 0.1582 | Yes |
| 15 | Col5a2 | 1443 | 0.396 | 0.1685 | Yes |
| 16 | Col5a1 | 1508 | 0.391 | 0.1767 | Yes |
| 17 | Col8a2 | 1511 | 0.390 | 0.1881 | Yes |
| 18 | Lrp1 | 1514 | 0.390 | 0.1995 | Yes |
| 19 | Tnfrsf12a | 1732 | 0.369 | 0.1992 | Yes |
| 20 | Rhob | 1779 | 0.364 | 0.2076 | Yes |
| 21 | Sdc4 | 1904 | 0.355 | 0.2117 | Yes |
| 22 | Col4a2 | 1983 | 0.349 | 0.2179 | Yes |
| 23 | Adam12 | 2043 | 0.344 | 0.2250 | Yes |
| 24 | Thbs1 | 2243 | 0.329 | 0.2245 | No |
| 25 | Sdc1 | 2654 | 0.301 | 0.2122 | No |
| 26 | Cald1 | 2921 | 0.284 | 0.2069 | No |
| 27 | Itgb3 | 3051 | 0.276 | 0.2083 | No |
| 28 | Plod1 | 3279 | 0.259 | 0.2043 | No |
| 29 | Itgav | 3461 | 0.248 | 0.2022 | No |
| 30 | Itgb1 | 3573 | 0.242 | 0.2036 | No |
| 31 | Vcam1 | 3604 | 0.240 | 0.2092 | No |
| 32 | Mmp2 | 4449 | 0.198 | 0.1714 | No |
| 33 | Comp | 4470 | 0.196 | 0.1762 | No |
| 34 | Sfrp4 | 4633 | 0.187 | 0.1734 | No |
| 35 | Tpm4 | 4711 | 0.184 | 0.1748 | No |
| 36 | Thbs2 | 4773 | 0.181 | 0.1770 | No |
| 37 | Sat1 | 5185 | 0.161 | 0.1606 | No |
| 38 | Magee1 | 5230 | 0.159 | 0.1630 | No |
| 39 | Itga5 | 5348 | 0.153 | 0.1615 | No |
| 40 | Mylk | 5448 | 0.148 | 0.1607 | No |
| 41 | Ppib | 5481 | 0.146 | 0.1634 | No |
| 42 | Lamc1 | 5653 | 0.139 | 0.1587 | No |
| 43 | Col1a2 | 5685 | 0.137 | 0.1611 | No |
| 44 | Plaur | 5748 | 0.135 | 0.1619 | No |
| 45 | Col1a1 | 5884 | 0.130 | 0.1587 | No |
| 46 | Thy1 | 5913 | 0.128 | 0.1611 | No |
| 47 | Basp1 | 6373 | 0.109 | 0.1406 | No |
| 48 | Plod2 | 6407 | 0.107 | 0.1421 | No |
| 49 | Fn1 | 6439 | 0.106 | 0.1436 | No |
| 50 | Ntm | 6445 | 0.106 | 0.1465 | No |
| 51 | Fstl1 | 6581 | 0.100 | 0.1424 | No |
| 52 | Id2 | 6593 | 0.100 | 0.1448 | No |
| 53 | Lama3 | 6749 | 0.093 | 0.1396 | No |
| 54 | Col5a3 | 6777 | 0.092 | 0.1409 | No |
| 55 | Calu | 7317 | 0.072 | 0.1152 | No |
| 56 | Nt5e | 7331 | 0.072 | 0.1166 | No |
| 57 | Slc6a8 | 7374 | 0.070 | 0.1165 | No |
| 58 | Lama1 | 7514 | 0.065 | 0.1113 | No |
| 59 | Fbn1 | 7844 | 0.053 | 0.0959 | No |
| 60 | Abi3bp | 7951 | 0.049 | 0.0918 | No |
| 61 | Flna | 7970 | 0.048 | 0.0923 | No |
| 62 | Sgcb | 8478 | 0.029 | 0.0670 | No |
| 63 | Ecm2 | 8563 | 0.025 | 0.0634 | No |
| 64 | Mmp14 | 9029 | 0.009 | 0.0397 | No |
| 65 | Cxcl12 | 9286 | 0.000 | 0.0265 | No |
| 66 | Plod3 | 9370 | -0.002 | 0.0223 | No |
| 67 | Col16a1 | 9423 | -0.004 | 0.0197 | No |
| 68 | Tnfaip3 | 9446 | -0.005 | 0.0187 | No |
| 69 | Pfn2 | 9778 | -0.017 | 0.0021 | No |
| 70 | Qsox1 | 9984 | -0.024 | -0.0077 | No |
| 71 | Cdh11 | 10153 | -0.030 | -0.0155 | No |
| 72 | Fbln1 | 10310 | -0.037 | -0.0225 | No |
| 73 | Cdh2 | 10355 | -0.039 | -0.0236 | No |
| 74 | Tpm1 | 10428 | -0.041 | -0.0261 | No |
| 75 | Postn | 10451 | -0.042 | -0.0260 | No |
| 76 | Igfbp4 | 10508 | -0.044 | -0.0276 | No |
| 77 | Dab2 | 10517 | -0.044 | -0.0267 | No |
| 78 | Gpx7 | 10779 | -0.053 | -0.0386 | No |
| 79 | Loxl1 | 10830 | -0.056 | -0.0395 | No |
| 80 | Col11a1 | 10940 | -0.060 | -0.0434 | No |
| 81 | Tpm2 | 10983 | -0.060 | -0.0438 | No |
| 82 | Fas | 10999 | -0.061 | -0.0427 | No |
| 83 | Tgfbr3 | 11082 | -0.064 | -0.0451 | No |
| 84 | Pmepa1 | 11245 | -0.071 | -0.0514 | No |
| 85 | Ecm1 | 11377 | -0.075 | -0.0559 | No |
| 86 | Capg | 11525 | -0.081 | -0.0611 | No |
| 87 | Fap | 11631 | -0.085 | -0.0640 | No |
| 88 | Edil3 | 11676 | -0.086 | -0.0638 | No |
| 89 | Wipf1 | 11678 | -0.087 | -0.0613 | No |
| 90 | Slit2 | 11989 | -0.100 | -0.0743 | No |
| 91 | Cdh6 | 12032 | -0.101 | -0.0735 | No |
| 92 | Notch2 | 12079 | -0.103 | -0.0728 | No |
| 93 | Vim | 12123 | -0.105 | -0.0720 | No |
| 94 | Vegfa | 12131 | -0.105 | -0.0692 | No |
| 95 | Tfpi2 | 12420 | -0.117 | -0.0806 | No |
| 96 | Bmp1 | 12874 | -0.135 | -0.1000 | No |
| 97 | Spock1 | 13044 | -0.142 | -0.1046 | No |
| 98 | Serpinh1 | 13386 | -0.156 | -0.1176 | No |
| 99 | Vegfc | 13543 | -0.163 | -0.1208 | No |
| 100 | Pvr | 13549 | -0.164 | -0.1162 | No |
| 101 | Gadd45a | 13667 | -0.169 | -0.1173 | No |
| 102 | Pdlim4 | 13671 | -0.169 | -0.1125 | No |
| 103 | Gadd45b | 13957 | -0.181 | -0.1218 | No |
| 104 | Eno2 | 14611 | -0.211 | -0.1493 | No |
| 105 | Pdgfrb | 14903 | -0.225 | -0.1577 | No |
| 106 | Il15 | 15200 | -0.240 | -0.1659 | No |
| 107 | Cd44 | 15214 | -0.241 | -0.1595 | No |
| 108 | Lgals1 | 15799 | -0.271 | -0.1816 | No |
| 109 | Sntb1 | 15917 | -0.278 | -0.1794 | No |
| 110 | Matn2 | 16153 | -0.293 | -0.1829 | No |
| 111 | Sgcd | 16273 | -0.301 | -0.1802 | No |
| 112 | Fzd8 | 16355 | -0.307 | -0.1753 | No |
| 113 | Emp3 | 16514 | -0.317 | -0.1741 | No |
| 114 | Spp1 | 16549 | -0.319 | -0.1664 | No |
| 115 | Serpine2 | 16612 | -0.323 | -0.1601 | No |
| 116 | Itga2 | 16664 | -0.327 | -0.1531 | No |
| 117 | Col7a1 | 16698 | -0.330 | -0.1451 | No |
| 118 | Fuca1 | 16898 | -0.343 | -0.1452 | No |
| 119 | Tgfb1 | 16902 | -0.344 | -0.1352 | No |
| 120 | Tagln | 16929 | -0.346 | -0.1264 | No |
| 121 | Colgalt1 | 16983 | -0.350 | -0.1188 | No |
| 122 | Fgf2 | 17073 | -0.355 | -0.1129 | No |
| 123 | Col12a1 | 17246 | -0.367 | -0.1109 | No |
| 124 | Nid2 | 17436 | -0.384 | -0.1094 | No |
| 125 | Dpysl3 | 17496 | -0.390 | -0.1009 | No |
| 126 | Cadm1 | 17523 | -0.393 | -0.0907 | No |
| 127 | Itgb5 | 17549 | -0.395 | -0.0803 | No |
| 128 | Gem | 17755 | -0.416 | -0.0786 | No |
| 129 | P3h1 | 17922 | -0.434 | -0.0744 | No |
| 130 | Timp3 | 17957 | -0.439 | -0.0632 | No |
| 131 | Gpc1 | 18000 | -0.444 | -0.0522 | No |
| 132 | Bgn | 18293 | -0.479 | -0.0532 | No |
| 133 | Pcolce | 18337 | -0.484 | -0.0411 | No |
| 134 | Fstl3 | 18461 | -0.502 | -0.0327 | No |
| 135 | Dcn | 18612 | -0.526 | -0.0249 | No |
| 136 | Anpep | 18614 | -0.527 | -0.0094 | No |
| 137 | Cap2 | 18704 | -0.543 | 0.0021 | No |
| 138 | Glipr1 | 18825 | -0.566 | 0.0126 | No |
| 139 | Lama2 | 19347 | -0.774 | 0.0085 | No |