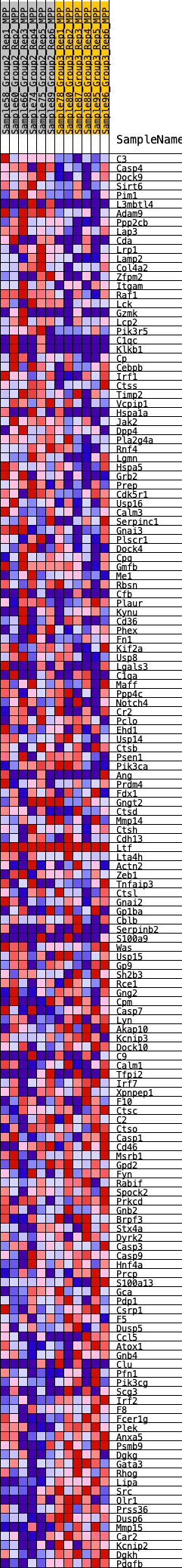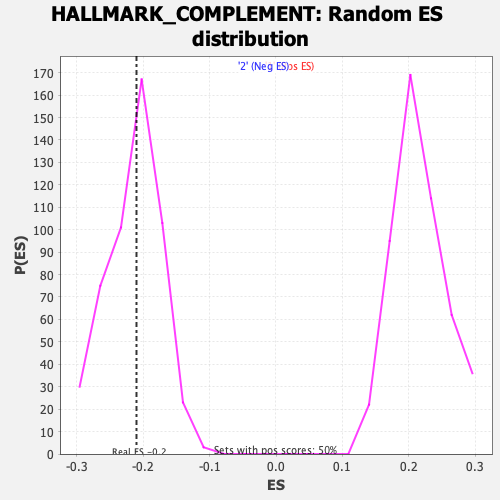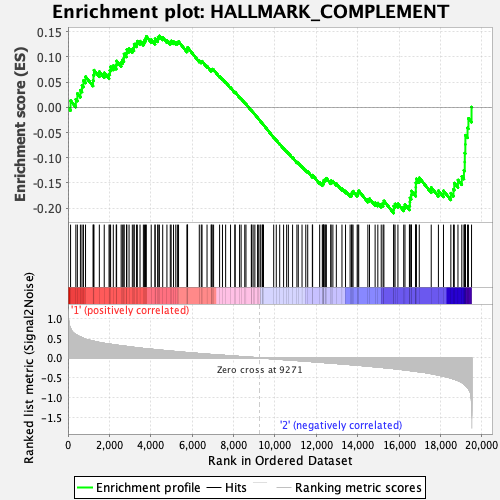
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.20990202 |
| Normalized Enrichment Score (NES) | -0.9815176 |
| Nominal p-value | 0.49800798 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | C3 | 124 | 0.744 | 0.0135 | No |
| 2 | Casp4 | 379 | 0.587 | 0.0161 | No |
| 3 | Dock9 | 452 | 0.567 | 0.0275 | No |
| 4 | Sirt6 | 599 | 0.528 | 0.0341 | No |
| 5 | Pim1 | 677 | 0.508 | 0.0438 | No |
| 6 | L3mbtl4 | 746 | 0.492 | 0.0534 | No |
| 7 | Adam9 | 847 | 0.473 | 0.0609 | No |
| 8 | Ppp2cb | 1210 | 0.424 | 0.0536 | No |
| 9 | Lap3 | 1232 | 0.422 | 0.0638 | No |
| 10 | Cda | 1255 | 0.420 | 0.0739 | No |
| 11 | Lrp1 | 1514 | 0.390 | 0.0710 | No |
| 12 | Lamp2 | 1751 | 0.367 | 0.0686 | No |
| 13 | Col4a2 | 1983 | 0.349 | 0.0660 | No |
| 14 | Zfpm2 | 2022 | 0.346 | 0.0733 | No |
| 15 | Itgam | 2058 | 0.343 | 0.0807 | No |
| 16 | Raf1 | 2189 | 0.334 | 0.0830 | No |
| 17 | Lck | 2330 | 0.323 | 0.0844 | No |
| 18 | Gzmk | 2349 | 0.321 | 0.0921 | No |
| 19 | Lcp2 | 2568 | 0.307 | 0.0890 | No |
| 20 | Pik3r5 | 2625 | 0.303 | 0.0942 | No |
| 21 | C1qc | 2702 | 0.298 | 0.0983 | No |
| 22 | Klkb1 | 2707 | 0.298 | 0.1061 | No |
| 23 | Cp | 2827 | 0.290 | 0.1077 | No |
| 24 | Cebpb | 2843 | 0.288 | 0.1146 | No |
| 25 | Irf1 | 2950 | 0.282 | 0.1167 | No |
| 26 | Ctss | 3108 | 0.271 | 0.1158 | No |
| 27 | Timp2 | 3186 | 0.266 | 0.1190 | No |
| 28 | Vcpip1 | 3196 | 0.265 | 0.1256 | No |
| 29 | Hspa1a | 3308 | 0.257 | 0.1267 | No |
| 30 | Jak2 | 3350 | 0.254 | 0.1314 | No |
| 31 | Dpp4 | 3480 | 0.247 | 0.1314 | No |
| 32 | Pla2g4a | 3642 | 0.238 | 0.1294 | No |
| 33 | Rnf4 | 3694 | 0.236 | 0.1331 | No |
| 34 | Lgmn | 3745 | 0.232 | 0.1367 | No |
| 35 | Hspa5 | 3784 | 0.230 | 0.1409 | No |
| 36 | Grb2 | 4021 | 0.219 | 0.1346 | No |
| 37 | Prep | 4205 | 0.210 | 0.1308 | No |
| 38 | Cdk5r1 | 4207 | 0.210 | 0.1363 | No |
| 39 | Usp16 | 4333 | 0.203 | 0.1353 | No |
| 40 | Calm3 | 4355 | 0.202 | 0.1396 | No |
| 41 | Serpinc1 | 4416 | 0.198 | 0.1418 | No |
| 42 | Gnai3 | 4576 | 0.190 | 0.1387 | No |
| 43 | Plscr1 | 4780 | 0.181 | 0.1331 | No |
| 44 | Dock4 | 4942 | 0.173 | 0.1294 | No |
| 45 | Cpq | 4984 | 0.171 | 0.1318 | No |
| 46 | Gmfb | 5099 | 0.165 | 0.1304 | No |
| 47 | Me1 | 5208 | 0.160 | 0.1291 | No |
| 48 | Rbsn | 5285 | 0.156 | 0.1293 | No |
| 49 | Cfb | 5341 | 0.153 | 0.1306 | No |
| 50 | Plaur | 5748 | 0.135 | 0.1132 | No |
| 51 | Kynu | 5769 | 0.134 | 0.1158 | No |
| 52 | Cd36 | 5777 | 0.134 | 0.1190 | No |
| 53 | Phex | 6344 | 0.110 | 0.0927 | No |
| 54 | Fn1 | 6439 | 0.106 | 0.0907 | No |
| 55 | Kif2a | 6476 | 0.104 | 0.0916 | No |
| 56 | Usp8 | 6718 | 0.095 | 0.0817 | No |
| 57 | Lgals3 | 6916 | 0.087 | 0.0738 | No |
| 58 | C1qa | 6944 | 0.086 | 0.0747 | No |
| 59 | Maff | 6965 | 0.085 | 0.0760 | No |
| 60 | Ppp4c | 7035 | 0.082 | 0.0746 | No |
| 61 | Notch4 | 7325 | 0.072 | 0.0616 | No |
| 62 | Cr2 | 7464 | 0.067 | 0.0563 | No |
| 63 | Pclo | 7617 | 0.062 | 0.0501 | No |
| 64 | Ehd1 | 7854 | 0.053 | 0.0393 | No |
| 65 | Usp14 | 8062 | 0.045 | 0.0298 | No |
| 66 | Ctsb | 8077 | 0.044 | 0.0302 | No |
| 67 | Psen1 | 8290 | 0.036 | 0.0202 | No |
| 68 | Pik3ca | 8365 | 0.033 | 0.0173 | No |
| 69 | Ang | 8541 | 0.026 | 0.0090 | No |
| 70 | Prdm4 | 8599 | 0.024 | 0.0066 | No |
| 71 | Fdx1 | 8858 | 0.015 | -0.0063 | No |
| 72 | Gngt2 | 8875 | 0.015 | -0.0067 | No |
| 73 | Ctsd | 8944 | 0.012 | -0.0099 | No |
| 74 | Mmp14 | 9029 | 0.009 | -0.0140 | No |
| 75 | Ctsh | 9160 | 0.004 | -0.0206 | No |
| 76 | Cdh13 | 9195 | 0.003 | -0.0223 | No |
| 77 | Ltf | 9279 | 0.000 | -0.0266 | No |
| 78 | Lta4h | 9319 | -0.001 | -0.0285 | No |
| 79 | Actn2 | 9397 | -0.003 | -0.0324 | No |
| 80 | Zeb1 | 9415 | -0.004 | -0.0332 | No |
| 81 | Tnfaip3 | 9446 | -0.005 | -0.0346 | No |
| 82 | Ctsl | 9939 | -0.023 | -0.0594 | No |
| 83 | Gnai2 | 10064 | -0.027 | -0.0651 | No |
| 84 | Gp1ba | 10229 | -0.033 | -0.0727 | No |
| 85 | Cblb | 10417 | -0.041 | -0.0813 | No |
| 86 | Serpinb2 | 10565 | -0.046 | -0.0876 | No |
| 87 | S100a9 | 10648 | -0.049 | -0.0905 | No |
| 88 | Was | 10852 | -0.057 | -0.0995 | No |
| 89 | Usp15 | 11061 | -0.064 | -0.1086 | No |
| 90 | Gp9 | 11127 | -0.066 | -0.1101 | No |
| 91 | Sh2b3 | 11308 | -0.073 | -0.1175 | No |
| 92 | Rce1 | 11489 | -0.079 | -0.1247 | No |
| 93 | Gng2 | 11583 | -0.083 | -0.1273 | No |
| 94 | Cpm | 11807 | -0.093 | -0.1363 | No |
| 95 | Casp7 | 11820 | -0.093 | -0.1344 | No |
| 96 | Lyn | 12160 | -0.106 | -0.1491 | No |
| 97 | Akap10 | 12288 | -0.112 | -0.1526 | No |
| 98 | Kcnip3 | 12317 | -0.112 | -0.1511 | No |
| 99 | Dock10 | 12348 | -0.114 | -0.1496 | No |
| 100 | C9 | 12355 | -0.114 | -0.1468 | No |
| 101 | Calm1 | 12366 | -0.115 | -0.1443 | No |
| 102 | Tfpi2 | 12420 | -0.117 | -0.1439 | No |
| 103 | Irf7 | 12455 | -0.118 | -0.1424 | No |
| 104 | Xpnpep1 | 12482 | -0.120 | -0.1406 | No |
| 105 | F10 | 12699 | -0.128 | -0.1483 | No |
| 106 | Ctsc | 12710 | -0.129 | -0.1454 | No |
| 107 | C2 | 12799 | -0.132 | -0.1464 | No |
| 108 | Ctso | 12966 | -0.139 | -0.1513 | No |
| 109 | Casp1 | 13238 | -0.149 | -0.1613 | No |
| 110 | Cd46 | 13410 | -0.157 | -0.1659 | No |
| 111 | Msrb1 | 13632 | -0.167 | -0.1728 | No |
| 112 | Gpd2 | 13703 | -0.170 | -0.1719 | No |
| 113 | Fyn | 13725 | -0.171 | -0.1684 | No |
| 114 | Rabif | 13773 | -0.173 | -0.1662 | No |
| 115 | Spock2 | 13973 | -0.182 | -0.1716 | No |
| 116 | Prkcd | 14012 | -0.183 | -0.1687 | No |
| 117 | Gnb2 | 14048 | -0.185 | -0.1655 | No |
| 118 | Brpf3 | 14489 | -0.206 | -0.1828 | No |
| 119 | Stx4a | 14568 | -0.209 | -0.1812 | No |
| 120 | Dyrk2 | 14840 | -0.222 | -0.1892 | No |
| 121 | Casp3 | 14971 | -0.228 | -0.1899 | No |
| 122 | Casp9 | 15135 | -0.237 | -0.1919 | No |
| 123 | Hnf4a | 15224 | -0.241 | -0.1900 | No |
| 124 | Prcp | 15264 | -0.244 | -0.1855 | No |
| 125 | S100a13 | 15737 | -0.268 | -0.2027 | Yes |
| 126 | Gca | 15742 | -0.269 | -0.1957 | Yes |
| 127 | Pdp1 | 15803 | -0.271 | -0.1916 | Yes |
| 128 | Csrp1 | 15942 | -0.280 | -0.1912 | Yes |
| 129 | F5 | 16221 | -0.297 | -0.1976 | Yes |
| 130 | Dusp5 | 16282 | -0.302 | -0.1926 | Yes |
| 131 | Ccl5 | 16497 | -0.315 | -0.1953 | Yes |
| 132 | Atox1 | 16519 | -0.317 | -0.1878 | Yes |
| 133 | Gnb4 | 16524 | -0.318 | -0.1795 | Yes |
| 134 | Clu | 16584 | -0.321 | -0.1740 | Yes |
| 135 | Pfn1 | 16595 | -0.322 | -0.1659 | Yes |
| 136 | Pik3cg | 16804 | -0.337 | -0.1676 | Yes |
| 137 | Scg3 | 16806 | -0.337 | -0.1586 | Yes |
| 138 | Irf2 | 16809 | -0.338 | -0.1497 | Yes |
| 139 | F8 | 16831 | -0.339 | -0.1417 | Yes |
| 140 | Fcer1g | 16973 | -0.349 | -0.1397 | Yes |
| 141 | Plek | 17551 | -0.396 | -0.1589 | Yes |
| 142 | Anxa5 | 17898 | -0.431 | -0.1652 | Yes |
| 143 | Psmb9 | 18143 | -0.461 | -0.1655 | Yes |
| 144 | Dgkg | 18499 | -0.508 | -0.1702 | Yes |
| 145 | Gata3 | 18625 | -0.529 | -0.1625 | Yes |
| 146 | Rhog | 18664 | -0.536 | -0.1501 | Yes |
| 147 | Lipa | 18843 | -0.569 | -0.1441 | Yes |
| 148 | Src | 19029 | -0.617 | -0.1371 | Yes |
| 149 | Olr1 | 19132 | -0.659 | -0.1248 | Yes |
| 150 | Prss36 | 19171 | -0.677 | -0.1086 | Yes |
| 151 | Dusp6 | 19174 | -0.677 | -0.0906 | Yes |
| 152 | Mmp15 | 19199 | -0.685 | -0.0735 | Yes |
| 153 | Car2 | 19202 | -0.687 | -0.0552 | Yes |
| 154 | Kcnip2 | 19306 | -0.750 | -0.0404 | Yes |
| 155 | Dgkh | 19349 | -0.776 | -0.0218 | Yes |
| 156 | Pdgfb | 19499 | -1.129 | 0.0007 | Yes |