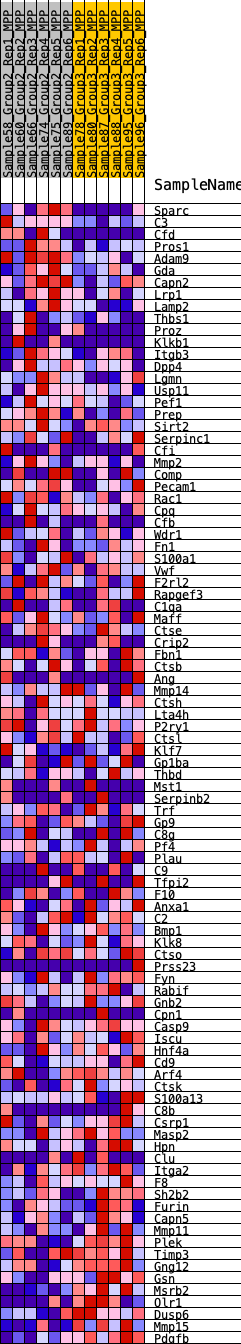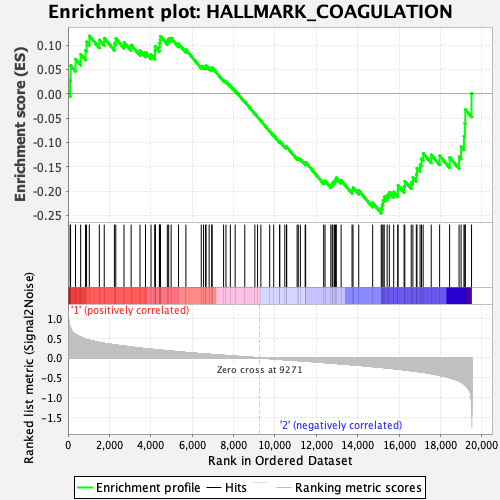
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.24552289 |
| Normalized Enrichment Score (NES) | -0.992012 |
| Nominal p-value | 0.47637796 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sparc | 104 | 0.766 | 0.0274 | No |
| 2 | C3 | 124 | 0.744 | 0.0583 | No |
| 3 | Cfd | 363 | 0.593 | 0.0714 | No |
| 4 | Pros1 | 611 | 0.526 | 0.0812 | No |
| 5 | Adam9 | 847 | 0.473 | 0.0894 | No |
| 6 | Gda | 897 | 0.465 | 0.1068 | No |
| 7 | Capn2 | 1030 | 0.447 | 0.1191 | No |
| 8 | Lrp1 | 1514 | 0.390 | 0.1110 | No |
| 9 | Lamp2 | 1751 | 0.367 | 0.1145 | No |
| 10 | Thbs1 | 2243 | 0.329 | 0.1033 | No |
| 11 | Proz | 2309 | 0.325 | 0.1139 | No |
| 12 | Klkb1 | 2707 | 0.298 | 0.1062 | No |
| 13 | Itgb3 | 3051 | 0.276 | 0.1003 | No |
| 14 | Dpp4 | 3480 | 0.247 | 0.0889 | No |
| 15 | Lgmn | 3745 | 0.232 | 0.0852 | No |
| 16 | Usp11 | 4011 | 0.219 | 0.0810 | No |
| 17 | Pef1 | 4203 | 0.210 | 0.0801 | No |
| 18 | Prep | 4205 | 0.210 | 0.0891 | No |
| 19 | Sirt2 | 4217 | 0.209 | 0.0975 | No |
| 20 | Serpinc1 | 4416 | 0.198 | 0.0958 | No |
| 21 | Cfi | 4421 | 0.198 | 0.1040 | No |
| 22 | Mmp2 | 4449 | 0.198 | 0.1111 | No |
| 23 | Comp | 4470 | 0.196 | 0.1185 | No |
| 24 | Pecam1 | 4800 | 0.180 | 0.1092 | No |
| 25 | Rac1 | 4864 | 0.176 | 0.1135 | No |
| 26 | Cpq | 4984 | 0.171 | 0.1148 | No |
| 27 | Cfb | 5341 | 0.153 | 0.1030 | No |
| 28 | Wdr1 | 5697 | 0.137 | 0.0905 | No |
| 29 | Fn1 | 6439 | 0.106 | 0.0569 | No |
| 30 | S100a1 | 6550 | 0.101 | 0.0556 | No |
| 31 | Vwf | 6649 | 0.098 | 0.0547 | No |
| 32 | F2rl2 | 6668 | 0.097 | 0.0579 | No |
| 33 | Rapgef3 | 6820 | 0.090 | 0.0540 | No |
| 34 | C1qa | 6944 | 0.086 | 0.0514 | No |
| 35 | Maff | 6965 | 0.085 | 0.0540 | No |
| 36 | Ctse | 7518 | 0.065 | 0.0283 | No |
| 37 | Crip2 | 7632 | 0.061 | 0.0251 | No |
| 38 | Fbn1 | 7844 | 0.053 | 0.0165 | No |
| 39 | Ctsb | 8077 | 0.044 | 0.0065 | No |
| 40 | Ang | 8541 | 0.026 | -0.0163 | No |
| 41 | Mmp14 | 9029 | 0.009 | -0.0410 | No |
| 42 | Ctsh | 9160 | 0.004 | -0.0475 | No |
| 43 | Lta4h | 9319 | -0.001 | -0.0556 | No |
| 44 | P2ry1 | 9746 | -0.016 | -0.0768 | No |
| 45 | Ctsl | 9939 | -0.023 | -0.0858 | No |
| 46 | Klf7 | 10228 | -0.033 | -0.0992 | No |
| 47 | Gp1ba | 10229 | -0.033 | -0.0977 | No |
| 48 | Thbd | 10476 | -0.043 | -0.1086 | No |
| 49 | Mst1 | 10562 | -0.046 | -0.1110 | No |
| 50 | Serpinb2 | 10565 | -0.046 | -0.1091 | No |
| 51 | Trf | 11070 | -0.064 | -0.1323 | No |
| 52 | Gp9 | 11127 | -0.066 | -0.1324 | No |
| 53 | C8g | 11228 | -0.070 | -0.1345 | No |
| 54 | Pf4 | 11460 | -0.079 | -0.1430 | No |
| 55 | Plau | 11483 | -0.079 | -0.1408 | No |
| 56 | C9 | 12355 | -0.114 | -0.1807 | No |
| 57 | Tfpi2 | 12420 | -0.117 | -0.1790 | No |
| 58 | F10 | 12699 | -0.128 | -0.1878 | No |
| 59 | Anxa1 | 12768 | -0.131 | -0.1857 | No |
| 60 | C2 | 12799 | -0.132 | -0.1816 | No |
| 61 | Bmp1 | 12874 | -0.135 | -0.1796 | No |
| 62 | Klk8 | 12924 | -0.138 | -0.1763 | No |
| 63 | Ctso | 12966 | -0.139 | -0.1724 | No |
| 64 | Prss23 | 13197 | -0.148 | -0.1779 | No |
| 65 | Fyn | 13725 | -0.171 | -0.1977 | No |
| 66 | Rabif | 13773 | -0.173 | -0.1927 | No |
| 67 | Gnb2 | 14048 | -0.185 | -0.1989 | No |
| 68 | Cpn1 | 14722 | -0.217 | -0.2243 | No |
| 69 | Casp9 | 15135 | -0.237 | -0.2354 | Yes |
| 70 | Iscu | 15178 | -0.238 | -0.2273 | Yes |
| 71 | Hnf4a | 15224 | -0.241 | -0.2193 | Yes |
| 72 | Cd9 | 15280 | -0.245 | -0.2117 | Yes |
| 73 | Arf4 | 15421 | -0.251 | -0.2081 | Yes |
| 74 | Ctsk | 15526 | -0.257 | -0.2025 | Yes |
| 75 | S100a13 | 15737 | -0.268 | -0.2018 | Yes |
| 76 | C8b | 15939 | -0.280 | -0.2002 | Yes |
| 77 | Csrp1 | 15942 | -0.280 | -0.1883 | Yes |
| 78 | Masp2 | 16239 | -0.299 | -0.1907 | Yes |
| 79 | Hpn | 16275 | -0.302 | -0.1796 | Yes |
| 80 | Clu | 16584 | -0.321 | -0.1817 | Yes |
| 81 | Itga2 | 16664 | -0.327 | -0.1718 | Yes |
| 82 | F8 | 16831 | -0.339 | -0.1658 | Yes |
| 83 | Sh2b2 | 16861 | -0.341 | -0.1527 | Yes |
| 84 | Furin | 17012 | -0.351 | -0.1454 | Yes |
| 85 | Capn5 | 17082 | -0.356 | -0.1338 | Yes |
| 86 | Mmp11 | 17161 | -0.361 | -0.1223 | Yes |
| 87 | Plek | 17551 | -0.396 | -0.1254 | Yes |
| 88 | Timp3 | 17957 | -0.439 | -0.1275 | Yes |
| 89 | Gng12 | 18440 | -0.499 | -0.1309 | Yes |
| 90 | Gsn | 18896 | -0.583 | -0.1294 | Yes |
| 91 | Msrb2 | 18994 | -0.606 | -0.1084 | Yes |
| 92 | Olr1 | 19132 | -0.659 | -0.0873 | Yes |
| 93 | Dusp6 | 19174 | -0.677 | -0.0604 | Yes |
| 94 | Mmp15 | 19199 | -0.685 | -0.0323 | Yes |
| 95 | Pdgfb | 19499 | -1.129 | 0.0007 | Yes |