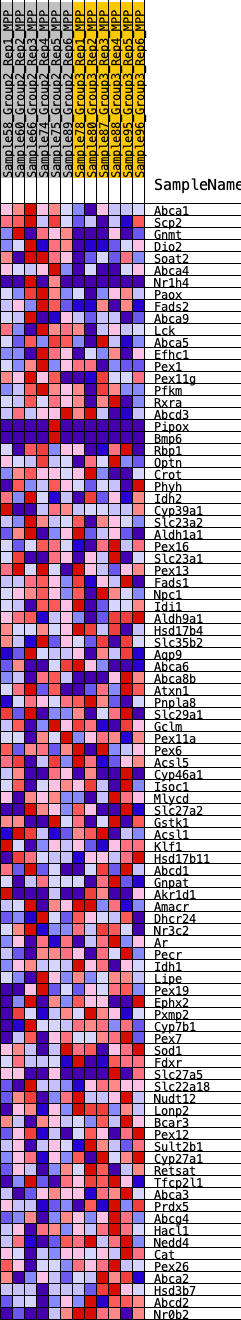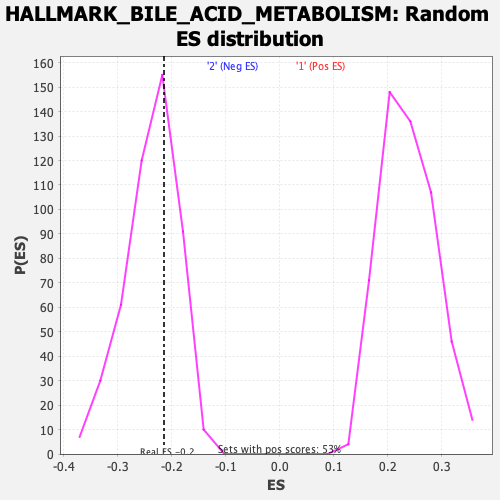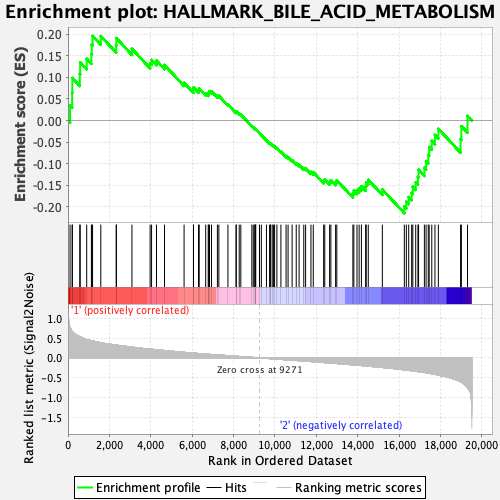
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group2_versus_Group3.MPP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.21427266 |
| Normalized Enrichment Score (NES) | -0.89840597 |
| Nominal p-value | 0.65189874 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 100 | 0.769 | 0.0350 | No |
| 2 | Scp2 | 202 | 0.663 | 0.0644 | No |
| 3 | Gnmt | 210 | 0.658 | 0.0984 | No |
| 4 | Dio2 | 570 | 0.535 | 0.1078 | No |
| 5 | Soat2 | 588 | 0.530 | 0.1346 | No |
| 6 | Abca4 | 904 | 0.465 | 0.1426 | No |
| 7 | Nr1h4 | 1130 | 0.434 | 0.1537 | No |
| 8 | Paox | 1146 | 0.432 | 0.1755 | No |
| 9 | Fads2 | 1187 | 0.428 | 0.1957 | No |
| 10 | Abca9 | 1585 | 0.383 | 0.1953 | No |
| 11 | Lck | 2330 | 0.323 | 0.1739 | No |
| 12 | Abca5 | 2338 | 0.323 | 0.1903 | No |
| 13 | Efhc1 | 3090 | 0.273 | 0.1659 | No |
| 14 | Pex1 | 3967 | 0.220 | 0.1323 | No |
| 15 | Pex11g | 4037 | 0.218 | 0.1401 | No |
| 16 | Pfkm | 4275 | 0.206 | 0.1386 | No |
| 17 | Rxra | 4664 | 0.187 | 0.1284 | No |
| 18 | Abcd3 | 5609 | 0.141 | 0.0871 | No |
| 19 | Pipox | 6061 | 0.122 | 0.0703 | No |
| 20 | Bmp6 | 6064 | 0.122 | 0.0765 | No |
| 21 | Rbp1 | 6307 | 0.112 | 0.0699 | No |
| 22 | Optn | 6339 | 0.110 | 0.0740 | No |
| 23 | Crot | 6653 | 0.097 | 0.0630 | No |
| 24 | Phyh | 6775 | 0.092 | 0.0615 | No |
| 25 | Idh2 | 6798 | 0.091 | 0.0651 | No |
| 26 | Cyp39a1 | 6837 | 0.090 | 0.0679 | No |
| 27 | Slc23a2 | 6933 | 0.086 | 0.0675 | No |
| 28 | Aldh1a1 | 7219 | 0.076 | 0.0568 | No |
| 29 | Pex16 | 7289 | 0.073 | 0.0570 | No |
| 30 | Slc23a1 | 7728 | 0.057 | 0.0375 | No |
| 31 | Pex13 | 8121 | 0.042 | 0.0195 | No |
| 32 | Fads1 | 8131 | 0.042 | 0.0212 | No |
| 33 | Npc1 | 8273 | 0.036 | 0.0159 | No |
| 34 | Idi1 | 8351 | 0.033 | 0.0136 | No |
| 35 | Aldh9a1 | 8884 | 0.014 | -0.0130 | No |
| 36 | Hsd17b4 | 8953 | 0.012 | -0.0159 | No |
| 37 | Slc35b2 | 9035 | 0.009 | -0.0196 | No |
| 38 | Aqp9 | 9039 | 0.009 | -0.0193 | No |
| 39 | Abca6 | 9040 | 0.009 | -0.0188 | No |
| 40 | Abca8b | 9072 | 0.007 | -0.0201 | No |
| 41 | Atxn1 | 9253 | 0.001 | -0.0293 | No |
| 42 | Pnpla8 | 9342 | -0.001 | -0.0337 | No |
| 43 | Slc29a1 | 9589 | -0.010 | -0.0459 | No |
| 44 | Gclm | 9740 | -0.016 | -0.0528 | No |
| 45 | Pex11a | 9776 | -0.017 | -0.0537 | No |
| 46 | Pex6 | 9803 | -0.018 | -0.0541 | No |
| 47 | Acsl5 | 9897 | -0.021 | -0.0578 | No |
| 48 | Cyp46a1 | 9924 | -0.022 | -0.0580 | No |
| 49 | Isoc1 | 9982 | -0.024 | -0.0597 | No |
| 50 | Mlycd | 10094 | -0.028 | -0.0639 | No |
| 51 | Slc27a2 | 10288 | -0.036 | -0.0720 | No |
| 52 | Gstk1 | 10537 | -0.045 | -0.0824 | No |
| 53 | Acsl1 | 10634 | -0.048 | -0.0848 | No |
| 54 | Klf1 | 10829 | -0.056 | -0.0919 | No |
| 55 | Hsd17b11 | 11028 | -0.062 | -0.0988 | No |
| 56 | Abcd1 | 11166 | -0.067 | -0.1024 | No |
| 57 | Gnpat | 11393 | -0.076 | -0.1100 | No |
| 58 | Akr1d1 | 11481 | -0.079 | -0.1104 | No |
| 59 | Amacr | 11739 | -0.090 | -0.1189 | No |
| 60 | Dhcr24 | 11851 | -0.094 | -0.1197 | No |
| 61 | Nr3c2 | 12352 | -0.114 | -0.1395 | No |
| 62 | Ar | 12406 | -0.117 | -0.1361 | No |
| 63 | Pecr | 12642 | -0.126 | -0.1416 | No |
| 64 | Idh1 | 12701 | -0.128 | -0.1379 | No |
| 65 | Lipe | 12931 | -0.138 | -0.1425 | No |
| 66 | Pex19 | 12988 | -0.140 | -0.1381 | No |
| 67 | Ephx2 | 13766 | -0.173 | -0.1691 | No |
| 68 | Pxmp2 | 13812 | -0.174 | -0.1623 | No |
| 69 | Cyp7b1 | 13964 | -0.181 | -0.1607 | No |
| 70 | Pex7 | 14067 | -0.185 | -0.1562 | No |
| 71 | Sod1 | 14177 | -0.190 | -0.1519 | No |
| 72 | Fdxr | 14383 | -0.200 | -0.1521 | No |
| 73 | Slc27a5 | 14408 | -0.201 | -0.1428 | No |
| 74 | Slc22a18 | 14509 | -0.207 | -0.1371 | No |
| 75 | Nudt12 | 15189 | -0.239 | -0.1596 | No |
| 76 | Lonp2 | 16251 | -0.300 | -0.1986 | Yes |
| 77 | Bcar3 | 16348 | -0.307 | -0.1876 | Yes |
| 78 | Pex12 | 16463 | -0.313 | -0.1771 | Yes |
| 79 | Sult2b1 | 16605 | -0.323 | -0.1675 | Yes |
| 80 | Cyp27a1 | 16661 | -0.326 | -0.1533 | Yes |
| 81 | Retsat | 16801 | -0.337 | -0.1429 | Yes |
| 82 | Tfcp2l1 | 16909 | -0.344 | -0.1304 | Yes |
| 83 | Abca3 | 16940 | -0.347 | -0.1138 | Yes |
| 84 | Prdx5 | 17220 | -0.366 | -0.1091 | Yes |
| 85 | Abcg4 | 17302 | -0.372 | -0.0939 | Yes |
| 86 | Hacl1 | 17408 | -0.381 | -0.0794 | Yes |
| 87 | Nedd4 | 17451 | -0.385 | -0.0615 | Yes |
| 88 | Cat | 17578 | -0.400 | -0.0471 | Yes |
| 89 | Pex26 | 17731 | -0.414 | -0.0333 | Yes |
| 90 | Abca2 | 17895 | -0.431 | -0.0192 | Yes |
| 91 | Hsd3b7 | 18969 | -0.601 | -0.0431 | Yes |
| 92 | Abcd2 | 19007 | -0.610 | -0.0131 | Yes |
| 93 | Nr0b2 | 19304 | -0.749 | 0.0107 | Yes |