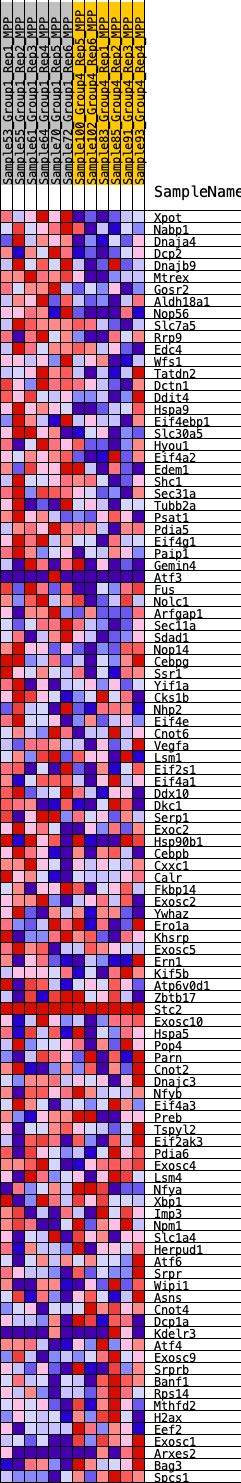
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
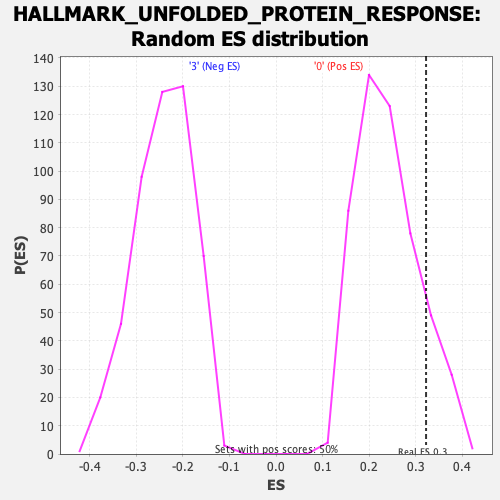
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.32222912 |
| Normalized Enrichment Score (NES) | 1.3411924 |
| Nominal p-value | 0.11904762 |
| FDR q-value | 0.33775482 |
| FWER p-Value | 0.762 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xpot | 346 | 0.640 | 0.0080 | Yes |
| 2 | Nabp1 | 348 | 0.639 | 0.0338 | Yes |
| 3 | Dnaja4 | 399 | 0.620 | 0.0563 | Yes |
| 4 | Dcp2 | 454 | 0.602 | 0.0778 | Yes |
| 5 | Dnajb9 | 700 | 0.545 | 0.0872 | Yes |
| 6 | Mtrex | 765 | 0.532 | 0.1054 | Yes |
| 7 | Gosr2 | 901 | 0.505 | 0.1188 | Yes |
| 8 | Aldh18a1 | 976 | 0.493 | 0.1349 | Yes |
| 9 | Nop56 | 1207 | 0.461 | 0.1417 | Yes |
| 10 | Slc7a5 | 1351 | 0.444 | 0.1523 | Yes |
| 11 | Rrp9 | 1415 | 0.436 | 0.1666 | Yes |
| 12 | Edc4 | 1627 | 0.414 | 0.1725 | Yes |
| 13 | Wfs1 | 1688 | 0.407 | 0.1858 | Yes |
| 14 | Tatdn2 | 1907 | 0.388 | 0.1903 | Yes |
| 15 | Dctn1 | 1974 | 0.381 | 0.2023 | Yes |
| 16 | Ddit4 | 2256 | 0.357 | 0.2022 | Yes |
| 17 | Hspa9 | 2311 | 0.352 | 0.2136 | Yes |
| 18 | Eif4ebp1 | 2386 | 0.347 | 0.2238 | Yes |
| 19 | Slc30a5 | 2431 | 0.343 | 0.2354 | Yes |
| 20 | Hyou1 | 2533 | 0.333 | 0.2436 | Yes |
| 21 | Eif4a2 | 2746 | 0.317 | 0.2455 | Yes |
| 22 | Edem1 | 2756 | 0.317 | 0.2579 | Yes |
| 23 | Shc1 | 2779 | 0.315 | 0.2695 | Yes |
| 24 | Sec31a | 2858 | 0.312 | 0.2780 | Yes |
| 25 | Tubb2a | 3216 | 0.291 | 0.2714 | Yes |
| 26 | Psat1 | 3330 | 0.283 | 0.2770 | Yes |
| 27 | Pdia5 | 3378 | 0.279 | 0.2859 | Yes |
| 28 | Eif4g1 | 3664 | 0.259 | 0.2816 | Yes |
| 29 | Paip1 | 4073 | 0.234 | 0.2701 | Yes |
| 30 | Gemin4 | 4077 | 0.234 | 0.2794 | Yes |
| 31 | Atf3 | 4081 | 0.234 | 0.2887 | Yes |
| 32 | Fus | 4185 | 0.229 | 0.2927 | Yes |
| 33 | Nolc1 | 4221 | 0.227 | 0.3000 | Yes |
| 34 | Arfgap1 | 4431 | 0.215 | 0.2980 | Yes |
| 35 | Sec11a | 4474 | 0.213 | 0.3044 | Yes |
| 36 | Sdad1 | 4523 | 0.210 | 0.3104 | Yes |
| 37 | Nop14 | 4535 | 0.210 | 0.3183 | Yes |
| 38 | Cebpg | 4621 | 0.206 | 0.3222 | Yes |
| 39 | Ssr1 | 5001 | 0.187 | 0.3102 | No |
| 40 | Yif1a | 5010 | 0.186 | 0.3173 | No |
| 41 | Cks1b | 5579 | 0.161 | 0.2946 | No |
| 42 | Nhp2 | 5760 | 0.152 | 0.2914 | No |
| 43 | Eif4e | 5897 | 0.145 | 0.2903 | No |
| 44 | Cnot6 | 6176 | 0.132 | 0.2813 | No |
| 45 | Vegfa | 6249 | 0.128 | 0.2828 | No |
| 46 | Lsm1 | 6327 | 0.125 | 0.2838 | No |
| 47 | Eif2s1 | 6354 | 0.123 | 0.2875 | No |
| 48 | Eif4a1 | 6668 | 0.112 | 0.2759 | No |
| 49 | Ddx10 | 6902 | 0.104 | 0.2680 | No |
| 50 | Dkc1 | 7043 | 0.097 | 0.2648 | No |
| 51 | Serp1 | 7101 | 0.095 | 0.2656 | No |
| 52 | Exoc2 | 7235 | 0.090 | 0.2624 | No |
| 53 | Hsp90b1 | 7424 | 0.082 | 0.2560 | No |
| 54 | Cebpb | 7459 | 0.080 | 0.2575 | No |
| 55 | Cxxc1 | 7702 | 0.069 | 0.2478 | No |
| 56 | Calr | 7738 | 0.067 | 0.2487 | No |
| 57 | Fkbp14 | 7918 | 0.059 | 0.2419 | No |
| 58 | Exosc2 | 7959 | 0.058 | 0.2421 | No |
| 59 | Ywhaz | 7982 | 0.057 | 0.2433 | No |
| 60 | Ero1a | 8343 | 0.043 | 0.2265 | No |
| 61 | Khsrp | 8390 | 0.040 | 0.2257 | No |
| 62 | Exosc5 | 8729 | 0.026 | 0.2094 | No |
| 63 | Ern1 | 8959 | 0.017 | 0.1982 | No |
| 64 | Kif5b | 8979 | 0.016 | 0.1979 | No |
| 65 | Atp6v0d1 | 9011 | 0.015 | 0.1969 | No |
| 66 | Zbtb17 | 9319 | 0.002 | 0.1811 | No |
| 67 | Stc2 | 9367 | 0.000 | 0.1787 | No |
| 68 | Exosc10 | 9431 | -0.000 | 0.1755 | No |
| 69 | Hspa5 | 9685 | -0.008 | 0.1628 | No |
| 70 | Pop4 | 9785 | -0.012 | 0.1582 | No |
| 71 | Parn | 10019 | -0.022 | 0.1470 | No |
| 72 | Cnot2 | 10236 | -0.030 | 0.1371 | No |
| 73 | Dnajc3 | 10319 | -0.033 | 0.1343 | No |
| 74 | Nfyb | 10637 | -0.046 | 0.1198 | No |
| 75 | Eif4a3 | 10850 | -0.054 | 0.1110 | No |
| 76 | Preb | 11086 | -0.064 | 0.1015 | No |
| 77 | Tspyl2 | 11112 | -0.065 | 0.1028 | No |
| 78 | Eif2ak3 | 11273 | -0.072 | 0.0975 | No |
| 79 | Pdia6 | 11274 | -0.072 | 0.1004 | No |
| 80 | Exosc4 | 11376 | -0.076 | 0.0983 | No |
| 81 | Lsm4 | 11702 | -0.090 | 0.0851 | No |
| 82 | Nfya | 12079 | -0.106 | 0.0701 | No |
| 83 | Xbp1 | 12642 | -0.133 | 0.0465 | No |
| 84 | Imp3 | 13042 | -0.149 | 0.0319 | No |
| 85 | Npm1 | 13239 | -0.159 | 0.0283 | No |
| 86 | Slc1a4 | 13327 | -0.164 | 0.0304 | No |
| 87 | Herpud1 | 13545 | -0.173 | 0.0262 | No |
| 88 | Atf6 | 13578 | -0.175 | 0.0316 | No |
| 89 | Srpr | 13898 | -0.188 | 0.0228 | No |
| 90 | Wipi1 | 13936 | -0.190 | 0.0286 | No |
| 91 | Asns | 14296 | -0.207 | 0.0184 | No |
| 92 | Cnot4 | 14383 | -0.211 | 0.0225 | No |
| 93 | Dcp1a | 14691 | -0.225 | 0.0158 | No |
| 94 | Kdelr3 | 15419 | -0.265 | -0.0110 | No |
| 95 | Atf4 | 15471 | -0.269 | -0.0027 | No |
| 96 | Exosc9 | 16614 | -0.342 | -0.0478 | No |
| 97 | Srprb | 16691 | -0.346 | -0.0377 | No |
| 98 | Banf1 | 17092 | -0.380 | -0.0430 | No |
| 99 | Rps14 | 17149 | -0.385 | -0.0303 | No |
| 100 | Mthfd2 | 17266 | -0.393 | -0.0204 | No |
| 101 | H2ax | 17337 | -0.400 | -0.0078 | No |
| 102 | Eef2 | 17408 | -0.406 | 0.0049 | No |
| 103 | Exosc1 | 18172 | -0.486 | -0.0148 | No |
| 104 | Arxes2 | 18564 | -0.543 | -0.0130 | No |
| 105 | Bag3 | 18946 | -0.622 | -0.0075 | No |
| 106 | Spcs1 | 19435 | -0.906 | 0.0040 | No |