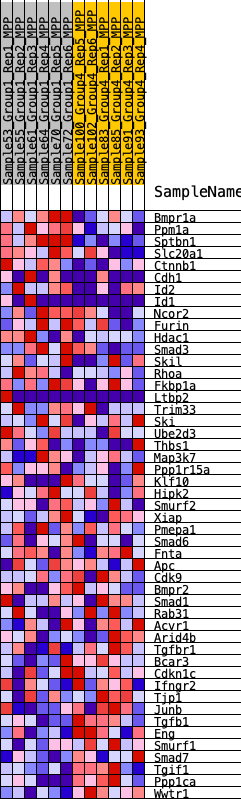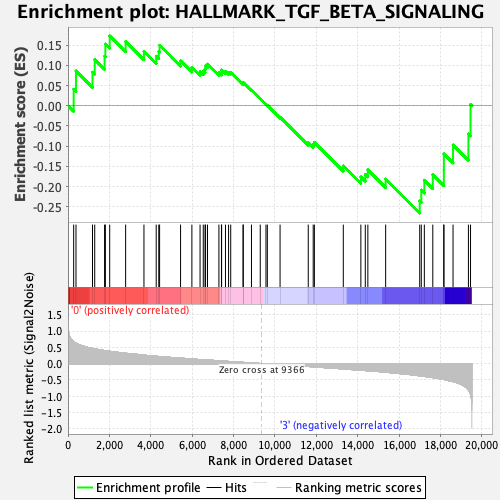
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.26590687 |
| Normalized Enrichment Score (NES) | -1.190692 |
| Nominal p-value | 0.23658052 |
| FDR q-value | 0.51010966 |
| FWER p-Value | 0.968 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmpr1a | 274 | 0.677 | 0.0411 | No |
| 2 | Ppm1a | 386 | 0.625 | 0.0863 | No |
| 3 | Sptbn1 | 1184 | 0.464 | 0.0832 | No |
| 4 | Slc20a1 | 1291 | 0.450 | 0.1145 | No |
| 5 | Ctnnb1 | 1767 | 0.399 | 0.1226 | No |
| 6 | Cdh1 | 1806 | 0.396 | 0.1530 | No |
| 7 | Id2 | 2015 | 0.377 | 0.1730 | No |
| 8 | Id1 | 2785 | 0.315 | 0.1592 | No |
| 9 | Ncor2 | 3670 | 0.259 | 0.1349 | No |
| 10 | Furin | 4262 | 0.224 | 0.1228 | No |
| 11 | Hdac1 | 4387 | 0.218 | 0.1342 | No |
| 12 | Smad3 | 4426 | 0.216 | 0.1498 | No |
| 13 | Skil | 5439 | 0.167 | 0.1114 | No |
| 14 | Rhoa | 5986 | 0.141 | 0.0949 | No |
| 15 | Fkbp1a | 6381 | 0.122 | 0.0846 | No |
| 16 | Ltbp2 | 6536 | 0.115 | 0.0861 | No |
| 17 | Trim33 | 6621 | 0.114 | 0.0911 | No |
| 18 | Ski | 6647 | 0.113 | 0.0990 | No |
| 19 | Ube2d3 | 6741 | 0.109 | 0.1031 | No |
| 20 | Thbs1 | 7292 | 0.087 | 0.0819 | No |
| 21 | Map3k7 | 7415 | 0.082 | 0.0824 | No |
| 22 | Ppp1r15a | 7423 | 0.082 | 0.0887 | No |
| 23 | Klf10 | 7609 | 0.074 | 0.0852 | No |
| 24 | Hipk2 | 7759 | 0.066 | 0.0829 | No |
| 25 | Smurf2 | 7862 | 0.061 | 0.0826 | No |
| 26 | Xiap | 8460 | 0.038 | 0.0550 | No |
| 27 | Pmepa1 | 8467 | 0.037 | 0.0577 | No |
| 28 | Smad6 | 8865 | 0.021 | 0.0390 | No |
| 29 | Fnta | 9289 | 0.003 | 0.0175 | No |
| 30 | Apc | 9566 | -0.004 | 0.0037 | No |
| 31 | Cdk9 | 9632 | -0.006 | 0.0009 | No |
| 32 | Bmpr2 | 10247 | -0.031 | -0.0282 | No |
| 33 | Smad1 | 11609 | -0.085 | -0.0911 | No |
| 34 | Rab31 | 11848 | -0.096 | -0.0955 | No |
| 35 | Acvr1 | 11905 | -0.099 | -0.0904 | No |
| 36 | Arid4b | 13302 | -0.162 | -0.1489 | No |
| 37 | Tgfbr1 | 14151 | -0.201 | -0.1761 | No |
| 38 | Bcar3 | 14365 | -0.211 | -0.1699 | No |
| 39 | Cdkn1c | 14489 | -0.217 | -0.1585 | No |
| 40 | Ifngr2 | 15349 | -0.261 | -0.1814 | No |
| 41 | Tjp1 | 16995 | -0.372 | -0.2356 | Yes |
| 42 | Junb | 17071 | -0.378 | -0.2086 | Yes |
| 43 | Tgfb1 | 17218 | -0.390 | -0.1843 | Yes |
| 44 | Eng | 17627 | -0.426 | -0.1705 | Yes |
| 45 | Smurf1 | 18161 | -0.484 | -0.1585 | Yes |
| 46 | Smad7 | 18162 | -0.484 | -0.1190 | Yes |
| 47 | Tgif1 | 18605 | -0.549 | -0.0970 | Yes |
| 48 | Ppp1ca | 19348 | -0.804 | -0.0697 | Yes |
| 49 | Wwtr1 | 19450 | -0.958 | 0.0032 | Yes |