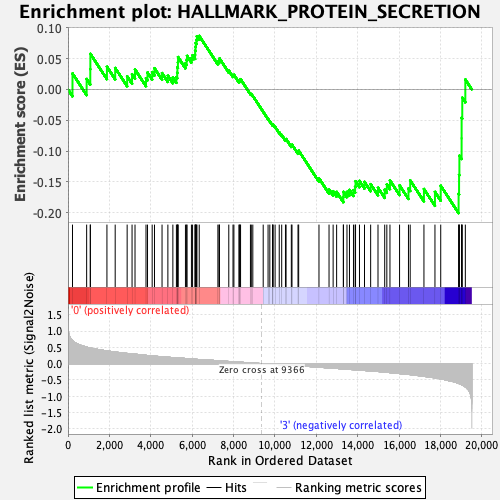
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
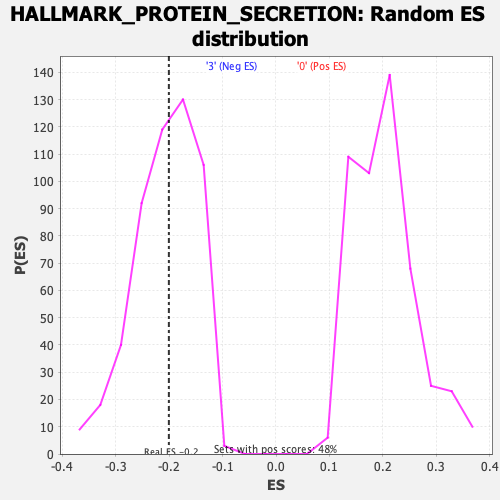
| Enrichment Score (ES) | -0.20068857 |
| Normalized Enrichment Score (NES) | -0.9752476 |
| Nominal p-value | 0.48162475 |
| FDR q-value | 0.8810254 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Igf2r | 217 | 0.712 | 0.0258 | No |
| 2 | Gosr2 | 901 | 0.505 | 0.0170 | No |
| 3 | Cltc | 1077 | 0.480 | 0.0329 | No |
| 4 | Arcn1 | 1081 | 0.480 | 0.0577 | No |
| 5 | Arfgef2 | 1881 | 0.389 | 0.0368 | No |
| 6 | Ykt6 | 2280 | 0.355 | 0.0348 | No |
| 7 | Sec31a | 2858 | 0.312 | 0.0213 | No |
| 8 | Yipf6 | 3095 | 0.298 | 0.0247 | No |
| 9 | Lman1 | 3238 | 0.290 | 0.0324 | No |
| 10 | Cav2 | 3769 | 0.253 | 0.0183 | No |
| 11 | Rab5a | 3842 | 0.248 | 0.0275 | No |
| 12 | Bnip3 | 4067 | 0.235 | 0.0282 | No |
| 13 | Scamp1 | 4179 | 0.230 | 0.0344 | No |
| 14 | Tmx1 | 4544 | 0.209 | 0.0265 | No |
| 15 | Sec22b | 4826 | 0.196 | 0.0223 | No |
| 16 | Ap2b1 | 5067 | 0.184 | 0.0195 | No |
| 17 | Stx16 | 5238 | 0.178 | 0.0200 | No |
| 18 | Ocrl | 5277 | 0.176 | 0.0272 | No |
| 19 | Atp1a1 | 5287 | 0.176 | 0.0359 | No |
| 20 | Atp6v1h | 5311 | 0.174 | 0.0438 | No |
| 21 | Arfip1 | 5318 | 0.174 | 0.0525 | No |
| 22 | Gla | 5675 | 0.157 | 0.0423 | No |
| 23 | Golga4 | 5732 | 0.153 | 0.0474 | No |
| 24 | Uso1 | 5751 | 0.152 | 0.0544 | No |
| 25 | Mapk1 | 5964 | 0.142 | 0.0509 | No |
| 26 | Arfgef1 | 6019 | 0.140 | 0.0554 | No |
| 27 | Tmed10 | 6140 | 0.134 | 0.0562 | No |
| 28 | Pam | 6144 | 0.134 | 0.0630 | No |
| 29 | Sh3gl2 | 6163 | 0.132 | 0.0690 | No |
| 30 | Tpd52 | 6173 | 0.132 | 0.0754 | No |
| 31 | Copb1 | 6212 | 0.130 | 0.0802 | No |
| 32 | Ap3b1 | 6228 | 0.130 | 0.0862 | No |
| 33 | Dst | 6341 | 0.124 | 0.0869 | No |
| 34 | Adam10 | 7243 | 0.089 | 0.0451 | No |
| 35 | Anp32e | 7297 | 0.087 | 0.0469 | No |
| 36 | M6pr | 7320 | 0.086 | 0.0502 | No |
| 37 | Napa | 7769 | 0.065 | 0.0306 | No |
| 38 | Snx2 | 7973 | 0.057 | 0.0231 | No |
| 39 | Vps45 | 8016 | 0.055 | 0.0238 | No |
| 40 | Lamp2 | 8258 | 0.046 | 0.0137 | No |
| 41 | Ica1 | 8296 | 0.044 | 0.0141 | No |
| 42 | Scrn1 | 8304 | 0.044 | 0.0160 | No |
| 43 | Vamp3 | 8338 | 0.043 | 0.0166 | No |
| 44 | Cog2 | 8815 | 0.023 | -0.0067 | No |
| 45 | Tsg101 | 8857 | 0.021 | -0.0077 | No |
| 46 | Rab14 | 8930 | 0.018 | -0.0105 | No |
| 47 | Tom1l1 | 9432 | -0.000 | -0.0363 | No |
| 48 | Vps4b | 9667 | -0.008 | -0.0480 | No |
| 49 | Rab22a | 9752 | -0.011 | -0.0517 | No |
| 50 | Zw10 | 9882 | -0.016 | -0.0575 | No |
| 51 | Gbf1 | 9902 | -0.017 | -0.0576 | No |
| 52 | Copb2 | 9905 | -0.017 | -0.0568 | No |
| 53 | Dop1a | 10007 | -0.022 | -0.0609 | No |
| 54 | Cope | 10207 | -0.029 | -0.0696 | No |
| 55 | Kif1b | 10326 | -0.034 | -0.0739 | No |
| 56 | Ctsc | 10508 | -0.041 | -0.0811 | No |
| 57 | Galc | 10537 | -0.042 | -0.0804 | No |
| 58 | Rab2a | 10791 | -0.051 | -0.0908 | No |
| 59 | Mon2 | 10823 | -0.053 | -0.0896 | No |
| 60 | Clcn3 | 11114 | -0.065 | -0.1011 | No |
| 61 | Cln5 | 11146 | -0.066 | -0.0993 | No |
| 62 | Stx7 | 12127 | -0.108 | -0.1441 | No |
| 63 | Vamp4 | 12613 | -0.132 | -0.1622 | No |
| 64 | Ap1g1 | 12811 | -0.139 | -0.1652 | No |
| 65 | Dnm1l | 12980 | -0.146 | -0.1662 | No |
| 66 | Arf1 | 13305 | -0.163 | -0.1745 | Yes |
| 67 | Ap2m1 | 13309 | -0.163 | -0.1661 | Yes |
| 68 | Rps6ka3 | 13480 | -0.171 | -0.1660 | Yes |
| 69 | Stx12 | 13600 | -0.176 | -0.1630 | Yes |
| 70 | Ppt1 | 13794 | -0.184 | -0.1634 | Yes |
| 71 | Stam | 13874 | -0.187 | -0.1577 | Yes |
| 72 | Rab9 | 13894 | -0.188 | -0.1489 | Yes |
| 73 | Sgms1 | 14084 | -0.198 | -0.1483 | Yes |
| 74 | Cd63 | 14327 | -0.209 | -0.1500 | Yes |
| 75 | Sspn | 14624 | -0.222 | -0.1537 | Yes |
| 76 | Ergic3 | 14979 | -0.240 | -0.1594 | Yes |
| 77 | Sec24d | 15303 | -0.258 | -0.1626 | Yes |
| 78 | Gnas | 15406 | -0.264 | -0.1541 | Yes |
| 79 | Ap3s1 | 15557 | -0.275 | -0.1476 | Yes |
| 80 | Tspan8 | 16021 | -0.303 | -0.1556 | Yes |
| 81 | Snap23 | 16451 | -0.328 | -0.1607 | Yes |
| 82 | Sod1 | 16535 | -0.335 | -0.1475 | Yes |
| 83 | Napg | 17199 | -0.388 | -0.1615 | Yes |
| 84 | Bet1 | 17731 | -0.436 | -0.1662 | Yes |
| 85 | Scamp3 | 18010 | -0.466 | -0.1563 | Yes |
| 86 | Arfgap3 | 18874 | -0.604 | -0.1693 | Yes |
| 87 | Krt18 | 18897 | -0.611 | -0.1386 | Yes |
| 88 | Abca1 | 18912 | -0.613 | -0.1075 | Yes |
| 89 | Ap2s1 | 19019 | -0.645 | -0.0794 | Yes |
| 90 | Clta | 19020 | -0.645 | -0.0459 | Yes |
| 91 | Atp7a | 19051 | -0.656 | -0.0133 | Yes |
| 92 | Rer1 | 19199 | -0.711 | 0.0161 | Yes |