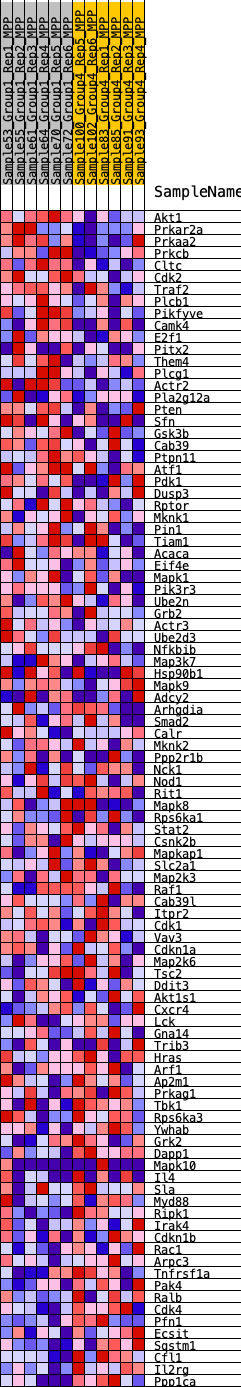
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
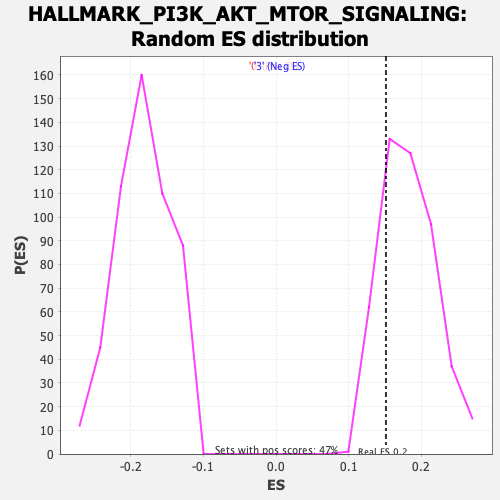
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.151443 |
| Normalized Enrichment Score (NES) | 0.8297576 |
| Nominal p-value | 0.779661 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 153 | 0.752 | 0.0267 | Yes |
| 2 | Prkar2a | 489 | 0.594 | 0.0368 | Yes |
| 3 | Prkaa2 | 491 | 0.593 | 0.0640 | Yes |
| 4 | Prkcb | 645 | 0.556 | 0.0817 | Yes |
| 5 | Cltc | 1077 | 0.480 | 0.0816 | Yes |
| 6 | Cdk2 | 1354 | 0.443 | 0.0878 | Yes |
| 7 | Traf2 | 1417 | 0.435 | 0.1047 | Yes |
| 8 | Plcb1 | 1464 | 0.432 | 0.1222 | Yes |
| 9 | Pikfyve | 1763 | 0.400 | 0.1252 | Yes |
| 10 | Camk4 | 1882 | 0.389 | 0.1371 | Yes |
| 11 | E2f1 | 2202 | 0.360 | 0.1372 | Yes |
| 12 | Pitx2 | 2454 | 0.340 | 0.1399 | Yes |
| 13 | Them4 | 2933 | 0.307 | 0.1294 | Yes |
| 14 | Plcg1 | 3279 | 0.287 | 0.1249 | Yes |
| 15 | Actr2 | 3498 | 0.272 | 0.1261 | Yes |
| 16 | Pla2g12a | 3608 | 0.264 | 0.1327 | Yes |
| 17 | Pten | 3925 | 0.243 | 0.1276 | Yes |
| 18 | Sfn | 4210 | 0.228 | 0.1234 | Yes |
| 19 | Gsk3b | 4461 | 0.214 | 0.1204 | Yes |
| 20 | Cab39 | 4600 | 0.207 | 0.1228 | Yes |
| 21 | Ptpn11 | 4798 | 0.198 | 0.1217 | Yes |
| 22 | Atf1 | 4845 | 0.195 | 0.1284 | Yes |
| 23 | Pdk1 | 4866 | 0.194 | 0.1363 | Yes |
| 24 | Dusp3 | 4943 | 0.190 | 0.1411 | Yes |
| 25 | Rptor | 4987 | 0.187 | 0.1475 | Yes |
| 26 | Mknk1 | 5080 | 0.183 | 0.1512 | Yes |
| 27 | Pin1 | 5405 | 0.169 | 0.1423 | Yes |
| 28 | Tiam1 | 5802 | 0.149 | 0.1288 | Yes |
| 29 | Acaca | 5846 | 0.148 | 0.1334 | Yes |
| 30 | Eif4e | 5897 | 0.145 | 0.1375 | Yes |
| 31 | Mapk1 | 5964 | 0.142 | 0.1406 | Yes |
| 32 | Pik3r3 | 5983 | 0.141 | 0.1462 | Yes |
| 33 | Ube2n | 6008 | 0.141 | 0.1514 | Yes |
| 34 | Grb2 | 6272 | 0.127 | 0.1437 | No |
| 35 | Actr3 | 6420 | 0.120 | 0.1417 | No |
| 36 | Ube2d3 | 6741 | 0.109 | 0.1302 | No |
| 37 | Nfkbib | 7156 | 0.093 | 0.1132 | No |
| 38 | Map3k7 | 7415 | 0.082 | 0.1036 | No |
| 39 | Hsp90b1 | 7424 | 0.082 | 0.1070 | No |
| 40 | Mapk9 | 7646 | 0.071 | 0.0989 | No |
| 41 | Adcy2 | 7659 | 0.071 | 0.1015 | No |
| 42 | Arhgdia | 7708 | 0.068 | 0.1022 | No |
| 43 | Smad2 | 7732 | 0.067 | 0.1041 | No |
| 44 | Calr | 7738 | 0.067 | 0.1069 | No |
| 45 | Mknk2 | 7925 | 0.059 | 0.1001 | No |
| 46 | Ppp2r1b | 8243 | 0.046 | 0.0858 | No |
| 47 | Nck1 | 8256 | 0.046 | 0.0873 | No |
| 48 | Nod1 | 8425 | 0.039 | 0.0804 | No |
| 49 | Rit1 | 8427 | 0.039 | 0.0822 | No |
| 50 | Mapk8 | 8711 | 0.027 | 0.0688 | No |
| 51 | Rps6ka1 | 8871 | 0.021 | 0.0616 | No |
| 52 | Stat2 | 8902 | 0.019 | 0.0610 | No |
| 53 | Csnk2b | 8918 | 0.019 | 0.0610 | No |
| 54 | Mapkap1 | 8991 | 0.015 | 0.0580 | No |
| 55 | Slc2a1 | 9018 | 0.014 | 0.0574 | No |
| 56 | Map2k3 | 9032 | 0.014 | 0.0573 | No |
| 57 | Raf1 | 9074 | 0.012 | 0.0558 | No |
| 58 | Cab39l | 9630 | -0.006 | 0.0275 | No |
| 59 | Itpr2 | 9782 | -0.012 | 0.0203 | No |
| 60 | Cdk1 | 10065 | -0.024 | 0.0068 | No |
| 61 | Vav3 | 10674 | -0.047 | -0.0223 | No |
| 62 | Cdkn1a | 10760 | -0.050 | -0.0244 | No |
| 63 | Map2k6 | 11059 | -0.063 | -0.0368 | No |
| 64 | Tsc2 | 11442 | -0.079 | -0.0529 | No |
| 65 | Ddit3 | 11627 | -0.086 | -0.0584 | No |
| 66 | Akt1s1 | 11923 | -0.099 | -0.0690 | No |
| 67 | Cxcr4 | 12295 | -0.116 | -0.0828 | No |
| 68 | Lck | 12511 | -0.127 | -0.0880 | No |
| 69 | Gna14 | 12805 | -0.139 | -0.0967 | No |
| 70 | Trib3 | 12985 | -0.146 | -0.0992 | No |
| 71 | Hras | 13079 | -0.151 | -0.0971 | No |
| 72 | Arf1 | 13305 | -0.163 | -0.1012 | No |
| 73 | Ap2m1 | 13309 | -0.163 | -0.0938 | No |
| 74 | Prkag1 | 13408 | -0.168 | -0.0912 | No |
| 75 | Tbk1 | 13469 | -0.170 | -0.0864 | No |
| 76 | Rps6ka3 | 13480 | -0.171 | -0.0791 | No |
| 77 | Ywhab | 13520 | -0.172 | -0.0732 | No |
| 78 | Grk2 | 14020 | -0.194 | -0.0900 | No |
| 79 | Dapp1 | 14062 | -0.196 | -0.0830 | No |
| 80 | Mapk10 | 14231 | -0.205 | -0.0822 | No |
| 81 | Il4 | 14480 | -0.216 | -0.0851 | No |
| 82 | Sla | 14702 | -0.226 | -0.0861 | No |
| 83 | Myd88 | 14858 | -0.233 | -0.0833 | No |
| 84 | Ripk1 | 14931 | -0.237 | -0.0761 | No |
| 85 | Irak4 | 14966 | -0.239 | -0.0669 | No |
| 86 | Cdkn1b | 15333 | -0.259 | -0.0738 | No |
| 87 | Rac1 | 16248 | -0.317 | -0.1063 | No |
| 88 | Arpc3 | 16668 | -0.345 | -0.1120 | No |
| 89 | Tnfrsf1a | 17244 | -0.392 | -0.1236 | No |
| 90 | Pak4 | 17268 | -0.394 | -0.1067 | No |
| 91 | Ralb | 17873 | -0.452 | -0.1170 | No |
| 92 | Cdk4 | 18083 | -0.474 | -0.1060 | No |
| 93 | Pfn1 | 18220 | -0.492 | -0.0903 | No |
| 94 | Ecsit | 18686 | -0.565 | -0.0883 | No |
| 95 | Sqstm1 | 19040 | -0.653 | -0.0764 | No |
| 96 | Cfl1 | 19160 | -0.691 | -0.0508 | No |
| 97 | Il2rg | 19162 | -0.691 | -0.0190 | No |
| 98 | Ppp1ca | 19348 | -0.804 | 0.0084 | No |