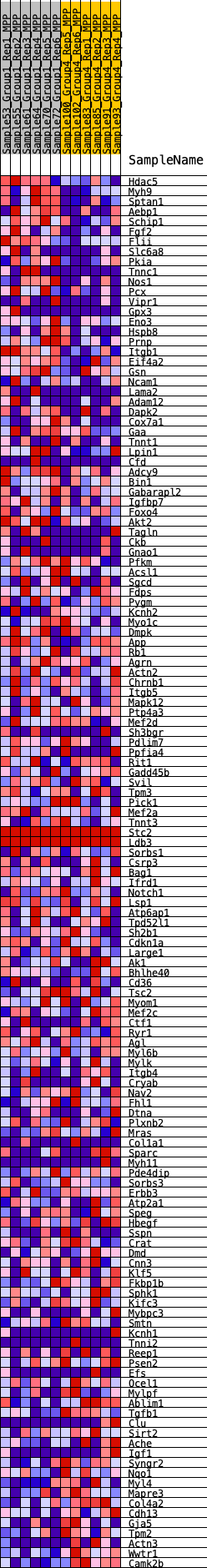
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
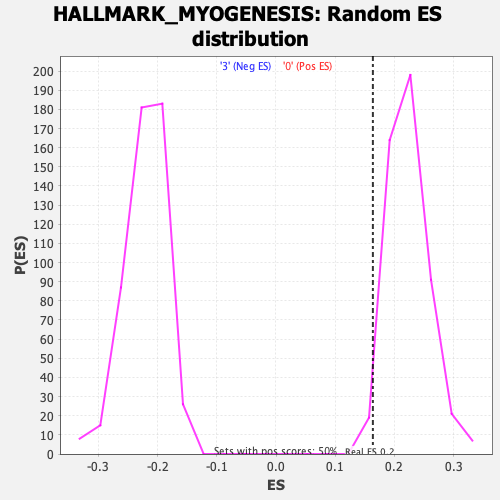
| Enrichment Score (ES) | 0.16369833 |
| Normalized Enrichment Score (NES) | 0.7331658 |
| Nominal p-value | 0.984 |
| FDR q-value | 0.94083667 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hdac5 | 259 | 0.687 | 0.0070 | Yes |
| 2 | Myh9 | 301 | 0.662 | 0.0245 | Yes |
| 3 | Sptan1 | 356 | 0.637 | 0.0406 | Yes |
| 4 | Aebp1 | 411 | 0.616 | 0.0561 | Yes |
| 5 | Schip1 | 478 | 0.596 | 0.0704 | Yes |
| 6 | Fgf2 | 611 | 0.564 | 0.0803 | Yes |
| 7 | Flii | 900 | 0.506 | 0.0804 | Yes |
| 8 | Slc6a8 | 928 | 0.500 | 0.0939 | Yes |
| 9 | Pkia | 996 | 0.490 | 0.1049 | Yes |
| 10 | Tnnc1 | 1012 | 0.489 | 0.1187 | Yes |
| 11 | Nos1 | 1185 | 0.464 | 0.1236 | Yes |
| 12 | Pcx | 1213 | 0.460 | 0.1358 | Yes |
| 13 | Vipr1 | 1725 | 0.403 | 0.1214 | Yes |
| 14 | Gpx3 | 1771 | 0.399 | 0.1309 | Yes |
| 15 | Eno3 | 1813 | 0.396 | 0.1406 | Yes |
| 16 | Hspb8 | 2309 | 0.353 | 0.1255 | Yes |
| 17 | Prnp | 2341 | 0.350 | 0.1342 | Yes |
| 18 | Itgb1 | 2545 | 0.331 | 0.1336 | Yes |
| 19 | Eif4a2 | 2746 | 0.317 | 0.1327 | Yes |
| 20 | Gsn | 2773 | 0.316 | 0.1407 | Yes |
| 21 | Ncam1 | 2941 | 0.307 | 0.1412 | Yes |
| 22 | Lama2 | 2946 | 0.306 | 0.1501 | Yes |
| 23 | Adam12 | 3248 | 0.289 | 0.1431 | Yes |
| 24 | Dapk2 | 3282 | 0.287 | 0.1499 | Yes |
| 25 | Cox7a1 | 3304 | 0.285 | 0.1573 | Yes |
| 26 | Gaa | 3446 | 0.276 | 0.1582 | Yes |
| 27 | Tnnt1 | 3609 | 0.264 | 0.1576 | Yes |
| 28 | Lpin1 | 4007 | 0.237 | 0.1442 | Yes |
| 29 | Cfd | 4041 | 0.236 | 0.1495 | Yes |
| 30 | Adcy9 | 4120 | 0.233 | 0.1524 | Yes |
| 31 | Bin1 | 4171 | 0.230 | 0.1566 | Yes |
| 32 | Gabarapl2 | 4247 | 0.225 | 0.1594 | Yes |
| 33 | Igfbp7 | 4293 | 0.222 | 0.1637 | Yes |
| 34 | Foxo4 | 4609 | 0.206 | 0.1536 | No |
| 35 | Akt2 | 4662 | 0.204 | 0.1569 | No |
| 36 | Tagln | 4785 | 0.199 | 0.1565 | No |
| 37 | Ckb | 5098 | 0.183 | 0.1458 | No |
| 38 | Gnao1 | 5125 | 0.182 | 0.1499 | No |
| 39 | Pfkm | 5253 | 0.178 | 0.1486 | No |
| 40 | Acsl1 | 5345 | 0.172 | 0.1490 | No |
| 41 | Sgcd | 5427 | 0.168 | 0.1498 | No |
| 42 | Fdps | 5748 | 0.152 | 0.1378 | No |
| 43 | Pygm | 5782 | 0.150 | 0.1406 | No |
| 44 | Kcnh2 | 5824 | 0.148 | 0.1429 | No |
| 45 | Myo1c | 6258 | 0.128 | 0.1243 | No |
| 46 | Dmpk | 6304 | 0.126 | 0.1257 | No |
| 47 | App | 6407 | 0.120 | 0.1240 | No |
| 48 | Rb1 | 6620 | 0.114 | 0.1165 | No |
| 49 | Agrn | 7131 | 0.094 | 0.0929 | No |
| 50 | Actn2 | 7286 | 0.087 | 0.0876 | No |
| 51 | Chrnb1 | 7319 | 0.086 | 0.0885 | No |
| 52 | Itgb5 | 7327 | 0.086 | 0.0907 | No |
| 53 | Mapk12 | 7376 | 0.084 | 0.0907 | No |
| 54 | Ptp4a3 | 7410 | 0.082 | 0.0914 | No |
| 55 | Mef2d | 7455 | 0.080 | 0.0915 | No |
| 56 | Sh3bgr | 7537 | 0.077 | 0.0896 | No |
| 57 | Pdlim7 | 8003 | 0.056 | 0.0673 | No |
| 58 | Ppfia4 | 8403 | 0.040 | 0.0478 | No |
| 59 | Rit1 | 8427 | 0.039 | 0.0478 | No |
| 60 | Gadd45b | 8650 | 0.029 | 0.0372 | No |
| 61 | Svil | 8724 | 0.026 | 0.0342 | No |
| 62 | Tpm3 | 8831 | 0.022 | 0.0294 | No |
| 63 | Pick1 | 8972 | 0.016 | 0.0227 | No |
| 64 | Mef2a | 9339 | 0.002 | 0.0038 | No |
| 65 | Tnnt3 | 9356 | 0.000 | 0.0030 | No |
| 66 | Stc2 | 9367 | 0.000 | 0.0025 | No |
| 67 | Ldb3 | 9368 | 0.000 | 0.0025 | No |
| 68 | Sorbs1 | 9589 | -0.005 | -0.0087 | No |
| 69 | Csrp3 | 9591 | -0.005 | -0.0086 | No |
| 70 | Bag1 | 9734 | -0.010 | -0.0157 | No |
| 71 | Ifrd1 | 9852 | -0.015 | -0.0213 | No |
| 72 | Notch1 | 9983 | -0.021 | -0.0274 | No |
| 73 | Lsp1 | 10176 | -0.028 | -0.0365 | No |
| 74 | Atp6ap1 | 10199 | -0.029 | -0.0367 | No |
| 75 | Tpd52l1 | 10291 | -0.033 | -0.0405 | No |
| 76 | Sh2b1 | 10739 | -0.050 | -0.0621 | No |
| 77 | Cdkn1a | 10760 | -0.050 | -0.0616 | No |
| 78 | Large1 | 10789 | -0.051 | -0.0615 | No |
| 79 | Ak1 | 10892 | -0.056 | -0.0651 | No |
| 80 | Bhlhe40 | 11048 | -0.062 | -0.0713 | No |
| 81 | Cd36 | 11241 | -0.071 | -0.0791 | No |
| 82 | Tsc2 | 11442 | -0.079 | -0.0871 | No |
| 83 | Myom1 | 11548 | -0.083 | -0.0900 | No |
| 84 | Mef2c | 11595 | -0.085 | -0.0899 | No |
| 85 | Ctf1 | 11778 | -0.093 | -0.0965 | No |
| 86 | Ryr1 | 11858 | -0.097 | -0.0977 | No |
| 87 | Agl | 12013 | -0.103 | -0.1026 | No |
| 88 | Myl6b | 12137 | -0.108 | -0.1058 | No |
| 89 | Mylk | 12203 | -0.111 | -0.1058 | No |
| 90 | Itgb4 | 12241 | -0.114 | -0.1044 | No |
| 91 | Cryab | 12355 | -0.119 | -0.1067 | No |
| 92 | Nav2 | 12371 | -0.120 | -0.1039 | No |
| 93 | Fhl1 | 12379 | -0.120 | -0.1007 | No |
| 94 | Dtna | 12401 | -0.121 | -0.0982 | No |
| 95 | Plxnb2 | 12446 | -0.124 | -0.0968 | No |
| 96 | Mras | 12862 | -0.141 | -0.1140 | No |
| 97 | Col1a1 | 12996 | -0.147 | -0.1165 | No |
| 98 | Sparc | 13594 | -0.176 | -0.1421 | No |
| 99 | Myh11 | 13686 | -0.180 | -0.1415 | No |
| 100 | Pde4dip | 13738 | -0.181 | -0.1387 | No |
| 101 | Sorbs3 | 13752 | -0.182 | -0.1340 | No |
| 102 | Erbb3 | 14075 | -0.197 | -0.1448 | No |
| 103 | Atp2a1 | 14160 | -0.201 | -0.1432 | No |
| 104 | Speg | 14288 | -0.206 | -0.1436 | No |
| 105 | Hbegf | 14481 | -0.216 | -0.1471 | No |
| 106 | Sspn | 14624 | -0.222 | -0.1478 | No |
| 107 | Crat | 14648 | -0.223 | -0.1424 | No |
| 108 | Dmd | 14810 | -0.231 | -0.1439 | No |
| 109 | Cnn3 | 14940 | -0.237 | -0.1435 | No |
| 110 | Klf5 | 15017 | -0.242 | -0.1402 | No |
| 111 | Fkbp1b | 15091 | -0.246 | -0.1367 | No |
| 112 | Sphk1 | 15276 | -0.256 | -0.1386 | No |
| 113 | Kifc3 | 15365 | -0.262 | -0.1353 | No |
| 114 | Mybpc3 | 15394 | -0.264 | -0.1290 | No |
| 115 | Smtn | 15650 | -0.281 | -0.1338 | No |
| 116 | Kcnh1 | 16127 | -0.309 | -0.1492 | No |
| 117 | Tnni2 | 16206 | -0.314 | -0.1439 | No |
| 118 | Reep1 | 16359 | -0.323 | -0.1422 | No |
| 119 | Psen2 | 16485 | -0.332 | -0.1388 | No |
| 120 | Efs | 16525 | -0.334 | -0.1309 | No |
| 121 | Ocel1 | 16660 | -0.344 | -0.1276 | No |
| 122 | Mylpf | 17016 | -0.374 | -0.1348 | No |
| 123 | Ablim1 | 17083 | -0.380 | -0.1270 | No |
| 124 | Tgfb1 | 17218 | -0.390 | -0.1223 | No |
| 125 | Clu | 17231 | -0.391 | -0.1113 | No |
| 126 | Sirt2 | 17377 | -0.403 | -0.1069 | No |
| 127 | Ache | 17399 | -0.405 | -0.0959 | No |
| 128 | Igf1 | 17531 | -0.416 | -0.0904 | No |
| 129 | Syngr2 | 17591 | -0.421 | -0.0809 | No |
| 130 | Nqo1 | 17693 | -0.431 | -0.0733 | No |
| 131 | Myl4 | 17796 | -0.443 | -0.0655 | No |
| 132 | Mapre3 | 17801 | -0.443 | -0.0525 | No |
| 133 | Col4a2 | 18093 | -0.474 | -0.0535 | No |
| 134 | Cdh13 | 18237 | -0.494 | -0.0462 | No |
| 135 | Gja5 | 18250 | -0.497 | -0.0321 | No |
| 136 | Tpm2 | 18640 | -0.556 | -0.0357 | No |
| 137 | Actn3 | 19027 | -0.649 | -0.0364 | No |
| 138 | Wwtr1 | 19450 | -0.958 | -0.0297 | No |
| 139 | Camk2b | 19497 | -1.109 | 0.0008 | No |