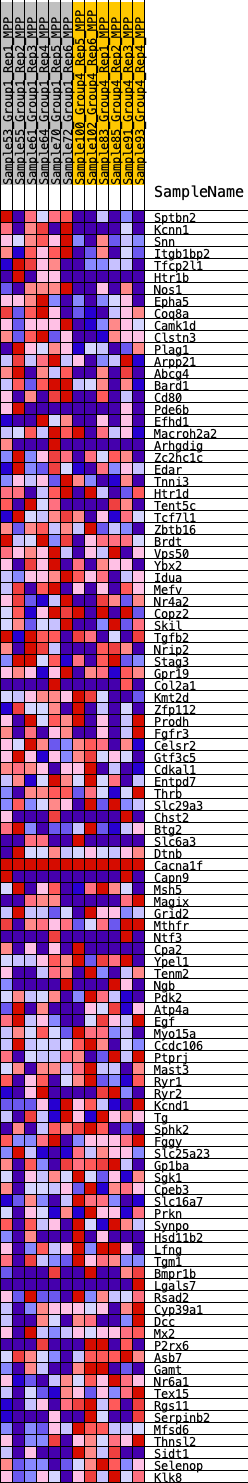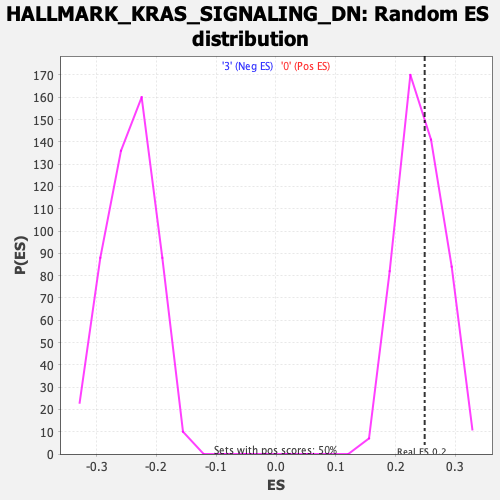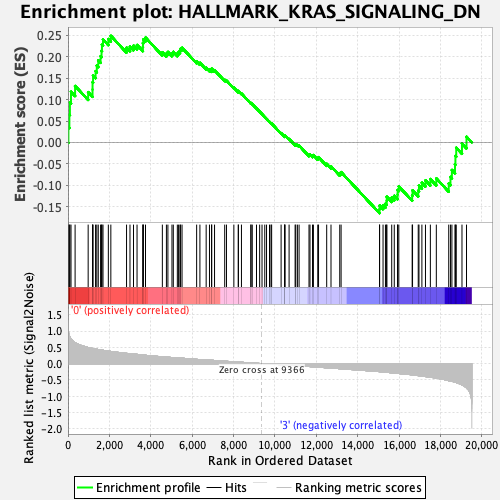
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.24862656 |
| Normalized Enrichment Score (NES) | 1.0259844 |
| Nominal p-value | 0.41616163 |
| FDR q-value | 0.8348345 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sptbn2 | 20 | 1.006 | 0.0358 | Yes |
| 2 | Kcnn1 | 66 | 0.856 | 0.0648 | Yes |
| 3 | Snn | 92 | 0.814 | 0.0933 | Yes |
| 4 | Itgb1bp2 | 145 | 0.757 | 0.1184 | Yes |
| 5 | Tfcp2l1 | 343 | 0.640 | 0.1316 | Yes |
| 6 | Htr1b | 974 | 0.493 | 0.1172 | Yes |
| 7 | Nos1 | 1185 | 0.464 | 0.1234 | Yes |
| 8 | Epha5 | 1187 | 0.464 | 0.1404 | Yes |
| 9 | Coq8a | 1211 | 0.460 | 0.1560 | Yes |
| 10 | Camk1d | 1332 | 0.446 | 0.1662 | Yes |
| 11 | Clstn3 | 1392 | 0.440 | 0.1792 | Yes |
| 12 | Plag1 | 1462 | 0.432 | 0.1915 | Yes |
| 13 | Arpp21 | 1574 | 0.419 | 0.2011 | Yes |
| 14 | Abcg4 | 1621 | 0.414 | 0.2139 | Yes |
| 15 | Bard1 | 1641 | 0.413 | 0.2280 | Yes |
| 16 | Cd80 | 1691 | 0.407 | 0.2404 | Yes |
| 17 | Pde6b | 1949 | 0.384 | 0.2412 | Yes |
| 18 | Efhd1 | 2070 | 0.372 | 0.2486 | Yes |
| 19 | Macroh2a2 | 2829 | 0.313 | 0.2210 | No |
| 20 | Arhgdig | 2995 | 0.305 | 0.2237 | No |
| 21 | Zc2hc1c | 3165 | 0.294 | 0.2258 | No |
| 22 | Edar | 3336 | 0.283 | 0.2273 | No |
| 23 | Tnni3 | 3618 | 0.263 | 0.2225 | No |
| 24 | Htr1d | 3622 | 0.263 | 0.2320 | No |
| 25 | Tent5c | 3642 | 0.261 | 0.2405 | No |
| 26 | Tcf7l1 | 3745 | 0.255 | 0.2446 | No |
| 27 | Zbtb16 | 4559 | 0.209 | 0.2103 | No |
| 28 | Brdt | 4755 | 0.200 | 0.2076 | No |
| 29 | Vps50 | 4827 | 0.196 | 0.2111 | No |
| 30 | Ybx2 | 5024 | 0.186 | 0.2078 | No |
| 31 | Idua | 5088 | 0.183 | 0.2113 | No |
| 32 | Mefv | 5283 | 0.176 | 0.2077 | No |
| 33 | Nr4a2 | 5343 | 0.173 | 0.2110 | No |
| 34 | Copz2 | 5409 | 0.169 | 0.2138 | No |
| 35 | Skil | 5439 | 0.167 | 0.2185 | No |
| 36 | Tgfb2 | 5510 | 0.164 | 0.2209 | No |
| 37 | Nrip2 | 6216 | 0.130 | 0.1893 | No |
| 38 | Stag3 | 6374 | 0.122 | 0.1857 | No |
| 39 | Gpr19 | 6676 | 0.112 | 0.1743 | No |
| 40 | Col2a1 | 6836 | 0.106 | 0.1700 | No |
| 41 | Kmt2d | 6940 | 0.102 | 0.1684 | No |
| 42 | Zfp112 | 6941 | 0.102 | 0.1721 | No |
| 43 | Prodh | 7081 | 0.095 | 0.1684 | No |
| 44 | Fgfr3 | 7572 | 0.075 | 0.1459 | No |
| 45 | Celsr2 | 7657 | 0.071 | 0.1442 | No |
| 46 | Gtf3c5 | 8014 | 0.055 | 0.1279 | No |
| 47 | Cdkal1 | 8228 | 0.047 | 0.1186 | No |
| 48 | Entpd7 | 8233 | 0.047 | 0.1201 | No |
| 49 | Thrb | 8381 | 0.041 | 0.1140 | No |
| 50 | Slc29a3 | 8835 | 0.022 | 0.0915 | No |
| 51 | Chst2 | 8843 | 0.022 | 0.0919 | No |
| 52 | Btg2 | 8903 | 0.019 | 0.0896 | No |
| 53 | Slc6a3 | 9108 | 0.011 | 0.0795 | No |
| 54 | Dtnb | 9261 | 0.004 | 0.0718 | No |
| 55 | Cacna1f | 9375 | 0.000 | 0.0660 | No |
| 56 | Capn9 | 9508 | -0.002 | 0.0593 | No |
| 57 | Msh5 | 9593 | -0.005 | 0.0552 | No |
| 58 | Magix | 9737 | -0.010 | 0.0482 | No |
| 59 | Grid2 | 9808 | -0.013 | 0.0450 | No |
| 60 | Mthfr | 9840 | -0.014 | 0.0440 | No |
| 61 | Ntf3 | 10297 | -0.033 | 0.0217 | No |
| 62 | Cpa2 | 10469 | -0.039 | 0.0143 | No |
| 63 | Ypel1 | 10472 | -0.039 | 0.0156 | No |
| 64 | Tenm2 | 10482 | -0.040 | 0.0166 | No |
| 65 | Ngb | 10683 | -0.047 | 0.0080 | No |
| 66 | Pdk2 | 10974 | -0.059 | -0.0047 | No |
| 67 | Atp4a | 11005 | -0.061 | -0.0041 | No |
| 68 | Egf | 11085 | -0.063 | -0.0058 | No |
| 69 | Myo15a | 11166 | -0.067 | -0.0075 | No |
| 70 | Ccdc106 | 11645 | -0.086 | -0.0289 | No |
| 71 | Ptprj | 11693 | -0.089 | -0.0281 | No |
| 72 | Mast3 | 11814 | -0.095 | -0.0308 | No |
| 73 | Ryr1 | 11858 | -0.097 | -0.0295 | No |
| 74 | Ryr2 | 12069 | -0.105 | -0.0365 | No |
| 75 | Kcnd1 | 12106 | -0.107 | -0.0344 | No |
| 76 | Tg | 12500 | -0.127 | -0.0500 | No |
| 77 | Sphk2 | 12708 | -0.137 | -0.0557 | No |
| 78 | Fggy | 13131 | -0.154 | -0.0718 | No |
| 79 | Slc25a23 | 13196 | -0.157 | -0.0693 | No |
| 80 | Gp1ba | 15053 | -0.244 | -0.1560 | No |
| 81 | Sgk1 | 15057 | -0.245 | -0.1472 | No |
| 82 | Cpeb3 | 15223 | -0.253 | -0.1464 | No |
| 83 | Slc16a7 | 15339 | -0.260 | -0.1428 | No |
| 84 | Prkn | 15400 | -0.264 | -0.1363 | No |
| 85 | Synpo | 15403 | -0.264 | -0.1267 | No |
| 86 | Hsd11b2 | 15642 | -0.280 | -0.1287 | No |
| 87 | Lfng | 15765 | -0.289 | -0.1244 | No |
| 88 | Tgm1 | 15925 | -0.298 | -0.1217 | No |
| 89 | Bmpr1b | 15928 | -0.298 | -0.1109 | No |
| 90 | Lgals7 | 15985 | -0.301 | -0.1028 | No |
| 91 | Rsad2 | 16630 | -0.342 | -0.1234 | No |
| 92 | Cyp39a1 | 16648 | -0.343 | -0.1117 | No |
| 93 | Dcc | 16923 | -0.365 | -0.1125 | No |
| 94 | Mx2 | 16959 | -0.368 | -0.1008 | No |
| 95 | P2rx6 | 17100 | -0.381 | -0.0941 | No |
| 96 | Asb7 | 17273 | -0.394 | -0.0885 | No |
| 97 | Gamt | 17509 | -0.415 | -0.0854 | No |
| 98 | Nr6a1 | 17797 | -0.443 | -0.0840 | No |
| 99 | Tex15 | 18397 | -0.522 | -0.0958 | No |
| 100 | Rgs11 | 18480 | -0.532 | -0.0805 | No |
| 101 | Serpinb2 | 18550 | -0.541 | -0.0643 | No |
| 102 | Mfsd6 | 18700 | -0.566 | -0.0512 | No |
| 103 | Thnsl2 | 18720 | -0.569 | -0.0314 | No |
| 104 | Sidt1 | 18760 | -0.577 | -0.0123 | No |
| 105 | Selenop | 19037 | -0.652 | -0.0026 | No |
| 106 | Klk8 | 19256 | -0.740 | 0.0132 | No |