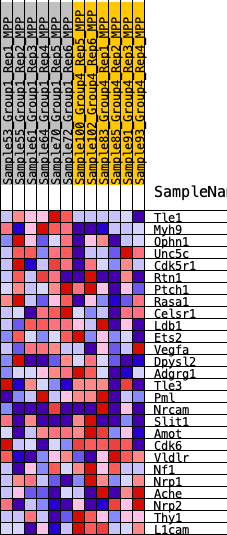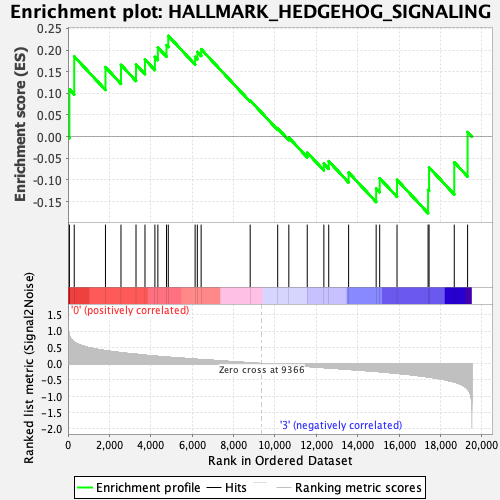
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.23286596 |
| Normalized Enrichment Score (NES) | 0.73068607 |
| Nominal p-value | 0.8353414 |
| FDR q-value | 0.88821745 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tle1 | 69 | 0.850 | 0.1092 | Yes |
| 2 | Myh9 | 301 | 0.662 | 0.1852 | Yes |
| 3 | Ophn1 | 1810 | 0.396 | 0.1604 | Yes |
| 4 | Unc5c | 2560 | 0.330 | 0.1658 | Yes |
| 5 | Cdk5r1 | 3286 | 0.287 | 0.1667 | Yes |
| 6 | Rtn1 | 3720 | 0.256 | 0.1784 | Yes |
| 7 | Ptch1 | 4199 | 0.229 | 0.1842 | Yes |
| 8 | Rasa1 | 4342 | 0.220 | 0.2062 | Yes |
| 9 | Celsr1 | 4760 | 0.200 | 0.2113 | Yes |
| 10 | Ldb1 | 4846 | 0.195 | 0.2329 | Yes |
| 11 | Ets2 | 6142 | 0.134 | 0.1842 | No |
| 12 | Vegfa | 6249 | 0.128 | 0.1958 | No |
| 13 | Dpysl2 | 6433 | 0.119 | 0.2021 | No |
| 14 | Adgrg1 | 8803 | 0.023 | 0.0837 | No |
| 15 | Tle3 | 10132 | -0.026 | 0.0190 | No |
| 16 | Pml | 10670 | -0.047 | -0.0024 | No |
| 17 | Nrcam | 11560 | -0.083 | -0.0369 | No |
| 18 | Slit1 | 12363 | -0.119 | -0.0623 | No |
| 19 | Amot | 12598 | -0.132 | -0.0568 | No |
| 20 | Cdk6 | 13559 | -0.174 | -0.0830 | No |
| 21 | Vldlr | 14890 | -0.235 | -0.1201 | No |
| 22 | Nf1 | 15061 | -0.245 | -0.0963 | No |
| 23 | Nrp1 | 15898 | -0.296 | -0.0999 | No |
| 24 | Ache | 17399 | -0.405 | -0.1231 | No |
| 25 | Nrp2 | 17447 | -0.409 | -0.0712 | No |
| 26 | Thy1 | 18664 | -0.560 | -0.0592 | No |
| 27 | L1cam | 19308 | -0.774 | 0.0105 | No |