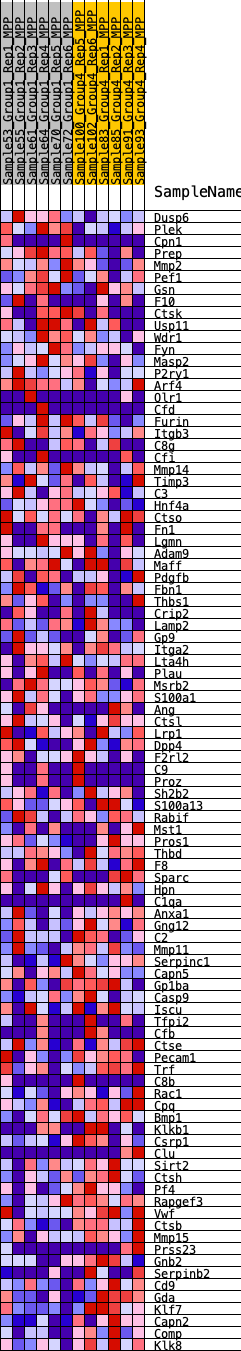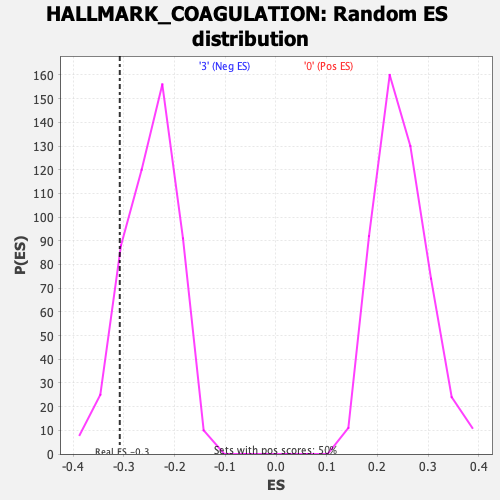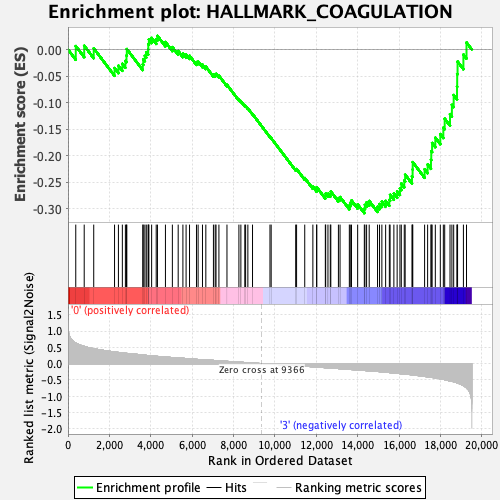
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.3080626 |
| Normalized Enrichment Score (NES) | -1.2353278 |
| Nominal p-value | 0.13052209 |
| FDR q-value | 0.59844625 |
| FWER p-Value | 0.936 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp6 | 375 | 0.629 | 0.0065 | No |
| 2 | Plek | 783 | 0.527 | 0.0072 | No |
| 3 | Cpn1 | 1243 | 0.456 | 0.0022 | No |
| 4 | Prep | 2249 | 0.357 | -0.0349 | No |
| 5 | Mmp2 | 2436 | 0.342 | -0.0304 | No |
| 6 | Pef1 | 2626 | 0.326 | -0.0268 | No |
| 7 | Gsn | 2773 | 0.316 | -0.0214 | No |
| 8 | F10 | 2826 | 0.314 | -0.0112 | No |
| 9 | Ctsk | 2842 | 0.313 | 0.0009 | No |
| 10 | Usp11 | 3613 | 0.263 | -0.0280 | No |
| 11 | Wdr1 | 3636 | 0.262 | -0.0184 | No |
| 12 | Fyn | 3714 | 0.256 | -0.0118 | No |
| 13 | Masp2 | 3783 | 0.251 | -0.0050 | No |
| 14 | P2ry1 | 3875 | 0.246 | 0.0004 | No |
| 15 | Arf4 | 3880 | 0.246 | 0.0103 | No |
| 16 | Olr1 | 3908 | 0.244 | 0.0189 | No |
| 17 | Cfd | 4041 | 0.236 | 0.0218 | No |
| 18 | Furin | 4262 | 0.224 | 0.0197 | No |
| 19 | Itgb3 | 4322 | 0.221 | 0.0257 | No |
| 20 | C8g | 4708 | 0.202 | 0.0142 | No |
| 21 | Cfi | 5043 | 0.185 | 0.0045 | No |
| 22 | Mmp14 | 5324 | 0.174 | -0.0028 | No |
| 23 | Timp3 | 5552 | 0.162 | -0.0078 | No |
| 24 | C3 | 5706 | 0.155 | -0.0093 | No |
| 25 | Hnf4a | 5872 | 0.146 | -0.0118 | No |
| 26 | Ctso | 6208 | 0.131 | -0.0237 | No |
| 27 | Fn1 | 6287 | 0.126 | -0.0225 | No |
| 28 | Lgmn | 6498 | 0.116 | -0.0286 | No |
| 29 | Adam9 | 6660 | 0.112 | -0.0323 | No |
| 30 | Maff | 7027 | 0.098 | -0.0471 | No |
| 31 | Pdgfb | 7110 | 0.094 | -0.0474 | No |
| 32 | Fbn1 | 7152 | 0.093 | -0.0457 | No |
| 33 | Thbs1 | 7292 | 0.087 | -0.0493 | No |
| 34 | Crip2 | 7681 | 0.070 | -0.0664 | No |
| 35 | Lamp2 | 8258 | 0.046 | -0.0942 | No |
| 36 | Gp9 | 8353 | 0.042 | -0.0973 | No |
| 37 | Itga2 | 8543 | 0.033 | -0.1057 | No |
| 38 | Lta4h | 8582 | 0.032 | -0.1064 | No |
| 39 | Plau | 8691 | 0.027 | -0.1108 | No |
| 40 | Msrb2 | 8916 | 0.019 | -0.1216 | No |
| 41 | S100a1 | 9759 | -0.011 | -0.1645 | No |
| 42 | Ang | 9817 | -0.013 | -0.1669 | No |
| 43 | Ctsl | 11002 | -0.060 | -0.2254 | No |
| 44 | Lrp1 | 11045 | -0.062 | -0.2250 | No |
| 45 | Dpp4 | 11439 | -0.079 | -0.2420 | No |
| 46 | F2rl2 | 11832 | -0.096 | -0.2583 | No |
| 47 | C9 | 12016 | -0.104 | -0.2634 | No |
| 48 | Proz | 12017 | -0.104 | -0.2592 | No |
| 49 | Sh2b2 | 12434 | -0.123 | -0.2755 | No |
| 50 | S100a13 | 12451 | -0.124 | -0.2712 | No |
| 51 | Rabif | 12561 | -0.130 | -0.2715 | No |
| 52 | Mst1 | 12673 | -0.135 | -0.2717 | No |
| 53 | Pros1 | 12699 | -0.137 | -0.2674 | No |
| 54 | Thbd | 13066 | -0.150 | -0.2801 | No |
| 55 | F8 | 13145 | -0.155 | -0.2777 | No |
| 56 | Sparc | 13594 | -0.176 | -0.2936 | No |
| 57 | Hpn | 13638 | -0.178 | -0.2885 | No |
| 58 | C1qa | 13698 | -0.180 | -0.2841 | No |
| 59 | Anxa1 | 13996 | -0.193 | -0.2915 | No |
| 60 | Gng12 | 14318 | -0.208 | -0.2995 | Yes |
| 61 | C2 | 14347 | -0.209 | -0.2924 | Yes |
| 62 | Mmp11 | 14428 | -0.214 | -0.2877 | Yes |
| 63 | Serpinc1 | 14554 | -0.220 | -0.2851 | Yes |
| 64 | Capn5 | 14957 | -0.239 | -0.2960 | Yes |
| 65 | Gp1ba | 15053 | -0.244 | -0.2909 | Yes |
| 66 | Casp9 | 15166 | -0.250 | -0.2864 | Yes |
| 67 | Iscu | 15348 | -0.261 | -0.2850 | Yes |
| 68 | Tfpi2 | 15527 | -0.272 | -0.2830 | Yes |
| 69 | Cfb | 15563 | -0.275 | -0.2736 | Yes |
| 70 | Ctse | 15746 | -0.287 | -0.2712 | Yes |
| 71 | Pecam1 | 15904 | -0.296 | -0.2671 | Yes |
| 72 | Trf | 16039 | -0.304 | -0.2615 | Yes |
| 73 | C8b | 16105 | -0.309 | -0.2522 | Yes |
| 74 | Rac1 | 16248 | -0.317 | -0.2465 | Yes |
| 75 | Cpq | 16291 | -0.320 | -0.2356 | Yes |
| 76 | Bmp1 | 16621 | -0.342 | -0.2385 | Yes |
| 77 | Klkb1 | 16651 | -0.343 | -0.2259 | Yes |
| 78 | Csrp1 | 16656 | -0.344 | -0.2120 | Yes |
| 79 | Clu | 17231 | -0.391 | -0.2255 | Yes |
| 80 | Sirt2 | 17377 | -0.403 | -0.2164 | Yes |
| 81 | Ctsh | 17539 | -0.416 | -0.2076 | Yes |
| 82 | Pf4 | 17553 | -0.417 | -0.1912 | Yes |
| 83 | Rapgef3 | 17594 | -0.422 | -0.1759 | Yes |
| 84 | Vwf | 17749 | -0.438 | -0.1659 | Yes |
| 85 | Ctsb | 17990 | -0.464 | -0.1593 | Yes |
| 86 | Mmp15 | 18136 | -0.480 | -0.1470 | Yes |
| 87 | Prss23 | 18202 | -0.491 | -0.1302 | Yes |
| 88 | Gnb2 | 18459 | -0.530 | -0.1217 | Yes |
| 89 | Serpinb2 | 18550 | -0.541 | -0.1041 | Yes |
| 90 | Cd9 | 18636 | -0.556 | -0.0857 | Yes |
| 91 | Gda | 18798 | -0.587 | -0.0699 | Yes |
| 92 | Klf7 | 18806 | -0.589 | -0.0461 | Yes |
| 93 | Capn2 | 18824 | -0.592 | -0.0227 | Yes |
| 94 | Comp | 19106 | -0.674 | -0.0095 | Yes |
| 95 | Klk8 | 19256 | -0.740 | 0.0132 | Yes |