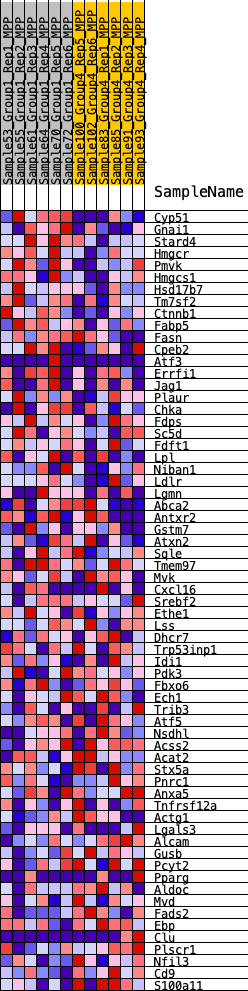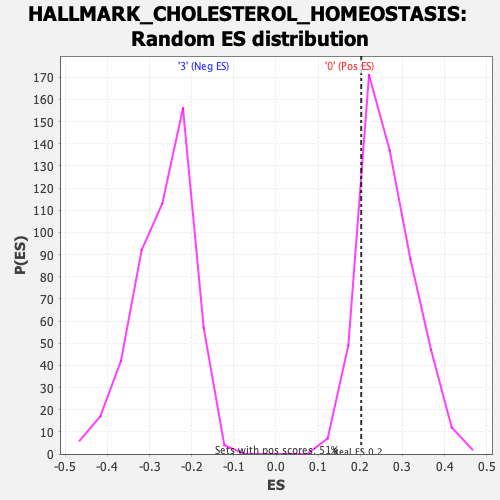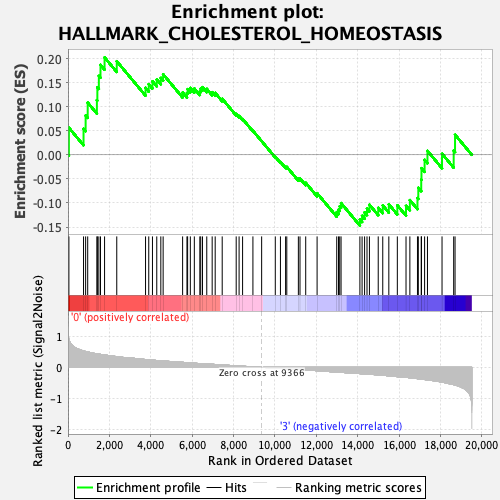
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group4.MPP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.20222129 |
| Normalized Enrichment Score (NES) | 0.76632434 |
| Nominal p-value | 0.83625734 |
| FDR q-value | 0.9583442 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 43 | 0.913 | 0.0561 | Yes |
| 2 | Gnai1 | 751 | 0.535 | 0.0540 | Yes |
| 3 | Stard4 | 852 | 0.514 | 0.0817 | Yes |
| 4 | Hmgcr | 952 | 0.496 | 0.1083 | Yes |
| 5 | Pmvk | 1396 | 0.440 | 0.1136 | Yes |
| 6 | Hmgcs1 | 1419 | 0.435 | 0.1403 | Yes |
| 7 | Hsd17b7 | 1491 | 0.428 | 0.1640 | Yes |
| 8 | Tm7sf2 | 1570 | 0.419 | 0.1868 | Yes |
| 9 | Ctnnb1 | 1767 | 0.399 | 0.2022 | Yes |
| 10 | Fabp5 | 2356 | 0.349 | 0.1943 | No |
| 11 | Fasn | 3747 | 0.255 | 0.1391 | No |
| 12 | Cpeb2 | 3902 | 0.245 | 0.1468 | No |
| 13 | Atf3 | 4081 | 0.234 | 0.1526 | No |
| 14 | Errfi1 | 4289 | 0.222 | 0.1562 | No |
| 15 | Jag1 | 4485 | 0.212 | 0.1597 | No |
| 16 | Plaur | 4597 | 0.207 | 0.1672 | No |
| 17 | Chka | 5538 | 0.163 | 0.1293 | No |
| 18 | Fdps | 5748 | 0.152 | 0.1283 | No |
| 19 | Sc5d | 5780 | 0.151 | 0.1363 | No |
| 20 | Fdft1 | 5911 | 0.144 | 0.1388 | No |
| 21 | Lpl | 6104 | 0.135 | 0.1376 | No |
| 22 | Niban1 | 6370 | 0.122 | 0.1318 | No |
| 23 | Ldlr | 6413 | 0.120 | 0.1373 | No |
| 24 | Lgmn | 6498 | 0.116 | 0.1404 | No |
| 25 | Abca2 | 6705 | 0.110 | 0.1369 | No |
| 26 | Antxr2 | 6966 | 0.101 | 0.1300 | No |
| 27 | Gstm7 | 7112 | 0.094 | 0.1285 | No |
| 28 | Atxn2 | 7451 | 0.081 | 0.1163 | No |
| 29 | Sqle | 8127 | 0.051 | 0.0849 | No |
| 30 | Tmem97 | 8267 | 0.045 | 0.0806 | No |
| 31 | Mvk | 8436 | 0.039 | 0.0744 | No |
| 32 | Cxcl16 | 8935 | 0.018 | 0.0499 | No |
| 33 | Srebf2 | 9357 | 0.000 | 0.0283 | No |
| 34 | Ethe1 | 10016 | -0.022 | -0.0041 | No |
| 35 | Lss | 10265 | -0.032 | -0.0148 | No |
| 36 | Dhcr7 | 10514 | -0.041 | -0.0250 | No |
| 37 | Trp53inp1 | 10572 | -0.043 | -0.0251 | No |
| 38 | Idi1 | 11131 | -0.066 | -0.0496 | No |
| 39 | Pdk3 | 11210 | -0.069 | -0.0492 | No |
| 40 | Fbxo6 | 11486 | -0.081 | -0.0582 | No |
| 41 | Ech1 | 12036 | -0.104 | -0.0797 | No |
| 42 | Trib3 | 12985 | -0.146 | -0.1192 | No |
| 43 | Atf5 | 13071 | -0.150 | -0.1139 | No |
| 44 | Nsdhl | 13129 | -0.154 | -0.1070 | No |
| 45 | Acss2 | 13194 | -0.157 | -0.1003 | No |
| 46 | Acat2 | 14104 | -0.198 | -0.1343 | No |
| 47 | Stx5a | 14210 | -0.204 | -0.1267 | No |
| 48 | Pnrc1 | 14331 | -0.209 | -0.1195 | No |
| 49 | Anxa5 | 14448 | -0.215 | -0.1118 | No |
| 50 | Tnfrsf12a | 14565 | -0.221 | -0.1036 | No |
| 51 | Actg1 | 14990 | -0.240 | -0.1101 | No |
| 52 | Lgals3 | 15209 | -0.253 | -0.1051 | No |
| 53 | Alcam | 15502 | -0.271 | -0.1028 | No |
| 54 | Gusb | 15913 | -0.297 | -0.1050 | No |
| 55 | Pcyt2 | 16333 | -0.322 | -0.1059 | No |
| 56 | Pparg | 16519 | -0.334 | -0.0941 | No |
| 57 | Aldoc | 16886 | -0.362 | -0.0897 | No |
| 58 | Mvd | 16927 | -0.366 | -0.0684 | No |
| 59 | Fads2 | 17068 | -0.378 | -0.0515 | No |
| 60 | Ebp | 17080 | -0.379 | -0.0278 | No |
| 61 | Clu | 17231 | -0.391 | -0.0105 | No |
| 62 | Plscr1 | 17369 | -0.403 | 0.0082 | No |
| 63 | Nfil3 | 18073 | -0.472 | 0.0022 | No |
| 64 | Cd9 | 18636 | -0.556 | 0.0088 | No |
| 65 | S100a11 | 18702 | -0.566 | 0.0416 | No |