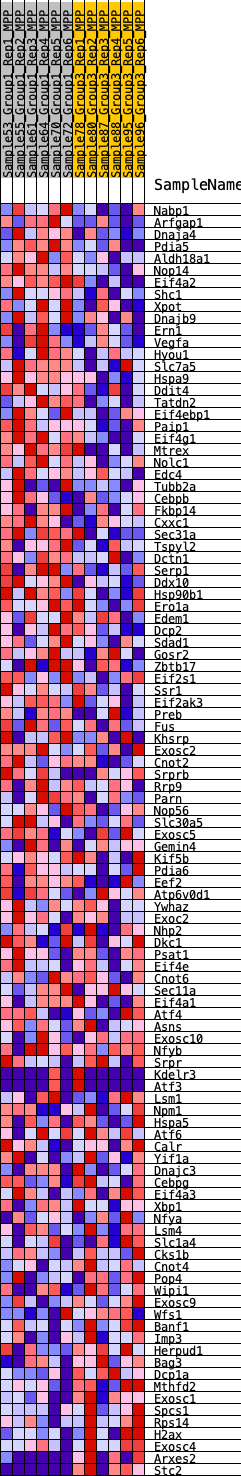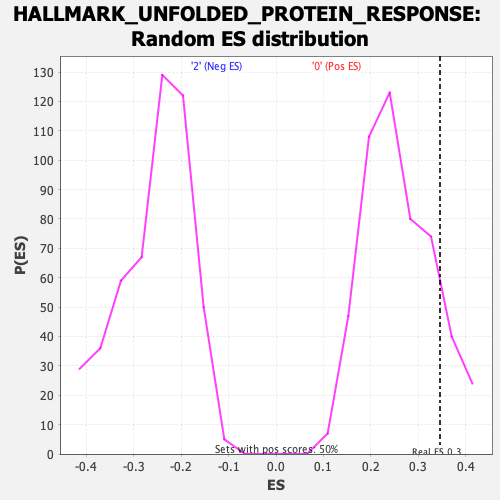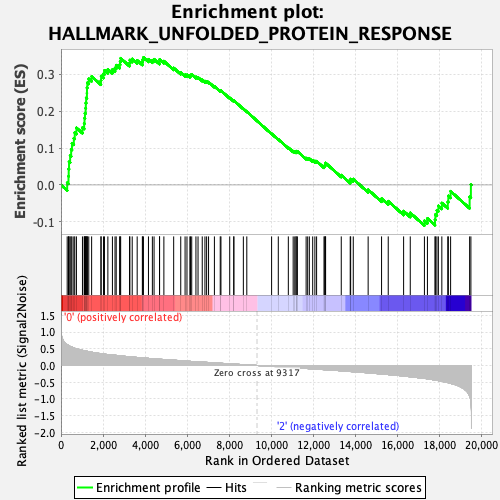
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group3.MPP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.34555414 |
| Normalized Enrichment Score (NES) | 1.3344886 |
| Nominal p-value | 0.13717695 |
| FDR q-value | 0.64673495 |
| FWER p-Value | 0.8 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nabp1 | 294 | 0.620 | 0.0061 | Yes |
| 2 | Arfgap1 | 356 | 0.598 | 0.0235 | Yes |
| 3 | Dnaja4 | 366 | 0.594 | 0.0434 | Yes |
| 4 | Pdia5 | 382 | 0.591 | 0.0629 | Yes |
| 5 | Aldh18a1 | 434 | 0.569 | 0.0797 | Yes |
| 6 | Nop14 | 482 | 0.554 | 0.0963 | Yes |
| 7 | Eif4a2 | 522 | 0.547 | 0.1130 | Yes |
| 8 | Shc1 | 610 | 0.523 | 0.1265 | Yes |
| 9 | Xpot | 654 | 0.512 | 0.1418 | Yes |
| 10 | Dnajb9 | 733 | 0.496 | 0.1548 | Yes |
| 11 | Ern1 | 1020 | 0.452 | 0.1556 | Yes |
| 12 | Vegfa | 1101 | 0.441 | 0.1666 | Yes |
| 13 | Hyou1 | 1118 | 0.439 | 0.1808 | Yes |
| 14 | Slc7a5 | 1139 | 0.435 | 0.1947 | Yes |
| 15 | Hspa9 | 1170 | 0.431 | 0.2079 | Yes |
| 16 | Ddit4 | 1180 | 0.430 | 0.2222 | Yes |
| 17 | Tatdn2 | 1197 | 0.428 | 0.2361 | Yes |
| 18 | Eif4ebp1 | 1232 | 0.424 | 0.2489 | Yes |
| 19 | Paip1 | 1235 | 0.424 | 0.2633 | Yes |
| 20 | Eif4g1 | 1251 | 0.421 | 0.2769 | Yes |
| 21 | Mtrex | 1320 | 0.410 | 0.2875 | Yes |
| 22 | Nolc1 | 1456 | 0.396 | 0.2941 | Yes |
| 23 | Edc4 | 1898 | 0.354 | 0.2836 | Yes |
| 24 | Tubb2a | 1907 | 0.354 | 0.2953 | Yes |
| 25 | Cebpb | 2019 | 0.342 | 0.3013 | Yes |
| 26 | Fkbp14 | 2066 | 0.338 | 0.3105 | Yes |
| 27 | Cxxc1 | 2228 | 0.324 | 0.3133 | Yes |
| 28 | Sec31a | 2444 | 0.311 | 0.3129 | Yes |
| 29 | Tspyl2 | 2568 | 0.305 | 0.3170 | Yes |
| 30 | Dctn1 | 2624 | 0.301 | 0.3245 | Yes |
| 31 | Serp1 | 2788 | 0.289 | 0.3260 | Yes |
| 32 | Ddx10 | 2810 | 0.288 | 0.3348 | Yes |
| 33 | Hsp90b1 | 2836 | 0.286 | 0.3433 | Yes |
| 34 | Ero1a | 3257 | 0.258 | 0.3306 | Yes |
| 35 | Edem1 | 3274 | 0.256 | 0.3385 | Yes |
| 36 | Dcp2 | 3385 | 0.251 | 0.3414 | Yes |
| 37 | Sdad1 | 3621 | 0.238 | 0.3375 | Yes |
| 38 | Gosr2 | 3867 | 0.227 | 0.3326 | Yes |
| 39 | Zbtb17 | 3882 | 0.226 | 0.3397 | Yes |
| 40 | Eif2s1 | 3918 | 0.224 | 0.3456 | Yes |
| 41 | Ssr1 | 4157 | 0.211 | 0.3405 | No |
| 42 | Eif2ak3 | 4343 | 0.202 | 0.3379 | No |
| 43 | Preb | 4424 | 0.198 | 0.3406 | No |
| 44 | Fus | 4684 | 0.186 | 0.3336 | No |
| 45 | Khsrp | 4685 | 0.186 | 0.3400 | No |
| 46 | Exosc2 | 4890 | 0.177 | 0.3356 | No |
| 47 | Cnot2 | 5356 | 0.158 | 0.3170 | No |
| 48 | Srprb | 5692 | 0.141 | 0.3046 | No |
| 49 | Rrp9 | 5898 | 0.132 | 0.2985 | No |
| 50 | Parn | 5988 | 0.127 | 0.2983 | No |
| 51 | Nop56 | 6126 | 0.122 | 0.2954 | No |
| 52 | Slc30a5 | 6157 | 0.121 | 0.2980 | No |
| 53 | Exosc5 | 6214 | 0.119 | 0.2992 | No |
| 54 | Gemin4 | 6414 | 0.113 | 0.2928 | No |
| 55 | Kif5b | 6514 | 0.110 | 0.2915 | No |
| 56 | Pdia6 | 6714 | 0.101 | 0.2847 | No |
| 57 | Eef2 | 6842 | 0.095 | 0.2814 | No |
| 58 | Atp6v0d1 | 6922 | 0.092 | 0.2805 | No |
| 59 | Ywhaz | 7014 | 0.088 | 0.2789 | No |
| 60 | Exoc2 | 7292 | 0.077 | 0.2672 | No |
| 61 | Nhp2 | 7569 | 0.066 | 0.2553 | No |
| 62 | Dkc1 | 7603 | 0.064 | 0.2557 | No |
| 63 | Psat1 | 8020 | 0.048 | 0.2360 | No |
| 64 | Eif4e | 8214 | 0.042 | 0.2275 | No |
| 65 | Cnot6 | 8215 | 0.042 | 0.2289 | No |
| 66 | Sec11a | 8668 | 0.024 | 0.2064 | No |
| 67 | Eif4a1 | 8832 | 0.018 | 0.1986 | No |
| 68 | Atf4 | 10011 | -0.024 | 0.1388 | No |
| 69 | Asns | 10329 | -0.037 | 0.1237 | No |
| 70 | Exosc10 | 10806 | -0.055 | 0.1011 | No |
| 71 | Nfyb | 11036 | -0.065 | 0.0915 | No |
| 72 | Srpr | 11113 | -0.068 | 0.0900 | No |
| 73 | Kdelr3 | 11175 | -0.071 | 0.0892 | No |
| 74 | Atf3 | 11177 | -0.071 | 0.0916 | No |
| 75 | Lsm1 | 11237 | -0.074 | 0.0911 | No |
| 76 | Npm1 | 11652 | -0.088 | 0.0728 | No |
| 77 | Hspa5 | 11726 | -0.091 | 0.0722 | No |
| 78 | Atf6 | 11805 | -0.094 | 0.0714 | No |
| 79 | Calr | 11964 | -0.100 | 0.0666 | No |
| 80 | Yif1a | 12068 | -0.104 | 0.0649 | No |
| 81 | Dnajc3 | 12150 | -0.107 | 0.0644 | No |
| 82 | Cebpg | 12502 | -0.122 | 0.0505 | No |
| 83 | Eif4a3 | 12536 | -0.124 | 0.0531 | No |
| 84 | Xbp1 | 12565 | -0.124 | 0.0559 | No |
| 85 | Nfya | 12578 | -0.125 | 0.0596 | No |
| 86 | Lsm4 | 13322 | -0.158 | 0.0267 | No |
| 87 | Slc1a4 | 13748 | -0.179 | 0.0109 | No |
| 88 | Cks1b | 13769 | -0.180 | 0.0160 | No |
| 89 | Cnot4 | 13893 | -0.185 | 0.0160 | No |
| 90 | Pop4 | 14600 | -0.218 | -0.0128 | No |
| 91 | Wipi1 | 15238 | -0.254 | -0.0370 | No |
| 92 | Exosc9 | 15555 | -0.270 | -0.0440 | No |
| 93 | Wfs1 | 16286 | -0.314 | -0.0708 | No |
| 94 | Banf1 | 16602 | -0.336 | -0.0755 | No |
| 95 | Imp3 | 17275 | -0.385 | -0.0969 | No |
| 96 | Herpud1 | 17418 | -0.397 | -0.0907 | No |
| 97 | Bag3 | 17778 | -0.434 | -0.0943 | No |
| 98 | Dcp1a | 17795 | -0.436 | -0.0801 | No |
| 99 | Mthfd2 | 17868 | -0.445 | -0.0686 | No |
| 100 | Exosc1 | 17938 | -0.451 | -0.0567 | No |
| 101 | Spcs1 | 18103 | -0.472 | -0.0489 | No |
| 102 | Rps14 | 18386 | -0.510 | -0.0460 | No |
| 103 | H2ax | 18412 | -0.512 | -0.0297 | No |
| 104 | Exosc4 | 18520 | -0.532 | -0.0170 | No |
| 105 | Arxes2 | 19424 | -0.911 | -0.0323 | No |
| 106 | Stc2 | 19484 | -1.071 | 0.0014 | No |