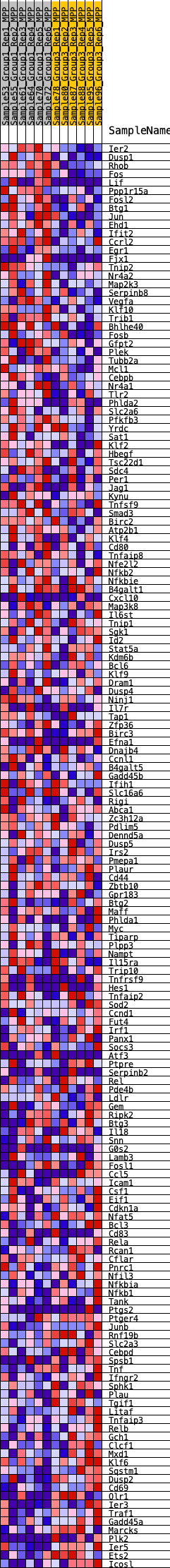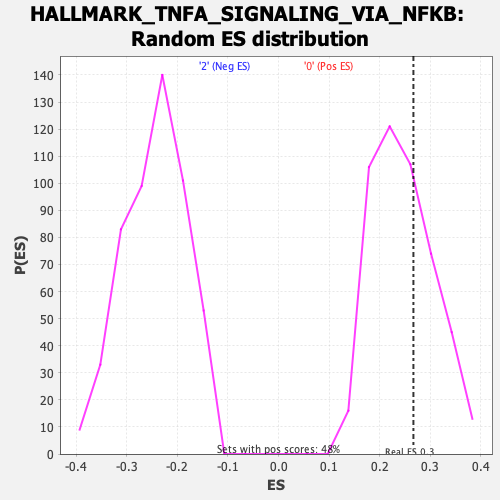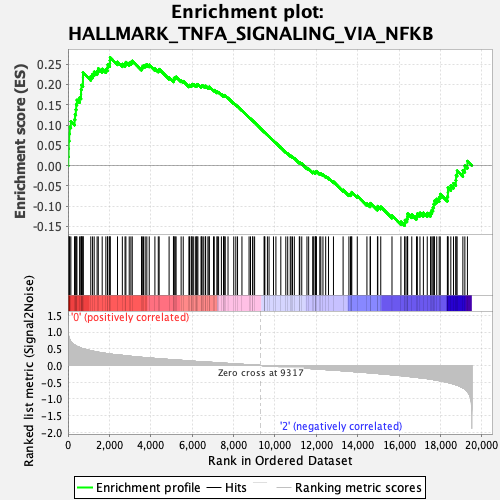
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group3.MPP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.26677448 |
| Normalized Enrichment Score (NES) | 1.0907805 |
| Nominal p-value | 0.32365146 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ier2 | 12 | 0.989 | 0.0221 | Yes |
| 2 | Dusp1 | 29 | 0.882 | 0.0416 | Yes |
| 3 | Rhob | 36 | 0.861 | 0.0610 | Yes |
| 4 | Fos | 58 | 0.814 | 0.0787 | Yes |
| 5 | Lif | 83 | 0.771 | 0.0951 | Yes |
| 6 | Ppp1r15a | 138 | 0.713 | 0.1087 | Yes |
| 7 | Fosl2 | 317 | 0.610 | 0.1136 | Yes |
| 8 | Btg1 | 341 | 0.603 | 0.1262 | Yes |
| 9 | Jun | 379 | 0.591 | 0.1379 | Yes |
| 10 | Ehd1 | 396 | 0.585 | 0.1505 | Yes |
| 11 | Ifit2 | 431 | 0.570 | 0.1618 | Yes |
| 12 | Ccrl2 | 561 | 0.537 | 0.1675 | Yes |
| 13 | Egr1 | 629 | 0.519 | 0.1760 | Yes |
| 14 | Fjx1 | 631 | 0.518 | 0.1878 | Yes |
| 15 | Tnip2 | 652 | 0.513 | 0.1986 | Yes |
| 16 | Nr4a2 | 722 | 0.499 | 0.2065 | Yes |
| 17 | Map2k3 | 723 | 0.498 | 0.2179 | Yes |
| 18 | Serpinb8 | 725 | 0.498 | 0.2293 | Yes |
| 19 | Vegfa | 1101 | 0.441 | 0.2201 | Yes |
| 20 | Klf10 | 1187 | 0.429 | 0.2255 | Yes |
| 21 | Trib1 | 1265 | 0.419 | 0.2312 | Yes |
| 22 | Bhlhe40 | 1406 | 0.401 | 0.2332 | Yes |
| 23 | Fosb | 1463 | 0.395 | 0.2394 | Yes |
| 24 | Gfpt2 | 1652 | 0.375 | 0.2383 | Yes |
| 25 | Plek | 1829 | 0.359 | 0.2374 | Yes |
| 26 | Tubb2a | 1907 | 0.354 | 0.2416 | Yes |
| 27 | Mcl1 | 1925 | 0.352 | 0.2488 | Yes |
| 28 | Cebpb | 2019 | 0.342 | 0.2518 | Yes |
| 29 | Nr4a1 | 2025 | 0.342 | 0.2594 | Yes |
| 30 | Tlr2 | 2035 | 0.341 | 0.2668 | Yes |
| 31 | Phlda2 | 2392 | 0.314 | 0.2556 | No |
| 32 | Slc2a6 | 2626 | 0.301 | 0.2505 | No |
| 33 | Pfkfb3 | 2756 | 0.292 | 0.2505 | No |
| 34 | Yrdc | 2797 | 0.289 | 0.2551 | No |
| 35 | Sat1 | 2958 | 0.277 | 0.2532 | No |
| 36 | Klf2 | 3039 | 0.272 | 0.2553 | No |
| 37 | Hbegf | 3105 | 0.268 | 0.2581 | No |
| 38 | Tsc22d1 | 3552 | 0.242 | 0.2406 | No |
| 39 | Sdc4 | 3590 | 0.240 | 0.2442 | No |
| 40 | Per1 | 3647 | 0.236 | 0.2468 | No |
| 41 | Jag1 | 3728 | 0.233 | 0.2480 | No |
| 42 | Kynu | 3809 | 0.229 | 0.2491 | No |
| 43 | Tnfsf9 | 3925 | 0.224 | 0.2483 | No |
| 44 | Smad3 | 4199 | 0.209 | 0.2390 | No |
| 45 | Birc2 | 4362 | 0.201 | 0.2353 | No |
| 46 | Atp2b1 | 4404 | 0.199 | 0.2377 | No |
| 47 | Klf4 | 4885 | 0.177 | 0.2170 | No |
| 48 | Cd80 | 5095 | 0.167 | 0.2100 | No |
| 49 | Tnfaip8 | 5115 | 0.167 | 0.2129 | No |
| 50 | Nfe2l2 | 5127 | 0.166 | 0.2161 | No |
| 51 | Nfkb2 | 5181 | 0.165 | 0.2172 | No |
| 52 | Nfkbie | 5223 | 0.163 | 0.2188 | No |
| 53 | B4galt1 | 5466 | 0.152 | 0.2098 | No |
| 54 | Cxcl10 | 5568 | 0.148 | 0.2080 | No |
| 55 | Map3k8 | 5843 | 0.134 | 0.1969 | No |
| 56 | Il6st | 5872 | 0.133 | 0.1985 | No |
| 57 | Tnip1 | 5953 | 0.129 | 0.1973 | No |
| 58 | Sgk1 | 5984 | 0.127 | 0.1987 | No |
| 59 | Id2 | 6000 | 0.127 | 0.2009 | No |
| 60 | Stat5a | 6074 | 0.124 | 0.1999 | No |
| 61 | Kdm6b | 6169 | 0.120 | 0.1978 | No |
| 62 | Bcl6 | 6223 | 0.118 | 0.1978 | No |
| 63 | Klf9 | 6226 | 0.118 | 0.2004 | No |
| 64 | Dram1 | 6276 | 0.116 | 0.2006 | No |
| 65 | Dusp4 | 6429 | 0.113 | 0.1953 | No |
| 66 | Ninj1 | 6458 | 0.111 | 0.1964 | No |
| 67 | Il7r | 6478 | 0.111 | 0.1980 | No |
| 68 | Tap1 | 6559 | 0.108 | 0.1963 | No |
| 69 | Zfp36 | 6640 | 0.105 | 0.1946 | No |
| 70 | Birc3 | 6651 | 0.104 | 0.1965 | No |
| 71 | Efna1 | 6761 | 0.099 | 0.1931 | No |
| 72 | Dnajb4 | 6818 | 0.096 | 0.1924 | No |
| 73 | Ccnl1 | 6826 | 0.096 | 0.1943 | No |
| 74 | B4galt5 | 7031 | 0.088 | 0.1857 | No |
| 75 | Gadd45b | 7066 | 0.086 | 0.1860 | No |
| 76 | Ifih1 | 7201 | 0.081 | 0.1809 | No |
| 77 | Slc16a6 | 7209 | 0.081 | 0.1824 | No |
| 78 | Rigi | 7276 | 0.078 | 0.1808 | No |
| 79 | Abca1 | 7412 | 0.072 | 0.1755 | No |
| 80 | Zc3h12a | 7514 | 0.068 | 0.1718 | No |
| 81 | Pdlim5 | 7536 | 0.067 | 0.1723 | No |
| 82 | Dennd5a | 7543 | 0.067 | 0.1735 | No |
| 83 | Dusp5 | 7598 | 0.064 | 0.1722 | No |
| 84 | Irs2 | 7732 | 0.060 | 0.1667 | No |
| 85 | Pmepa1 | 8009 | 0.049 | 0.1535 | No |
| 86 | Plaur | 8109 | 0.046 | 0.1495 | No |
| 87 | Cd44 | 8192 | 0.042 | 0.1462 | No |
| 88 | Zbtb10 | 8405 | 0.034 | 0.1360 | No |
| 89 | Gpr183 | 8749 | 0.021 | 0.1188 | No |
| 90 | Btg2 | 8818 | 0.019 | 0.1157 | No |
| 91 | Maff | 8921 | 0.015 | 0.1108 | No |
| 92 | Phlda1 | 8931 | 0.014 | 0.1106 | No |
| 93 | Myc | 9017 | 0.011 | 0.1065 | No |
| 94 | Tiparp | 9478 | -0.005 | 0.0828 | No |
| 95 | Plpp3 | 9490 | -0.005 | 0.0824 | No |
| 96 | Nampt | 9513 | -0.006 | 0.0814 | No |
| 97 | Il15ra | 9636 | -0.010 | 0.0753 | No |
| 98 | Trip10 | 9715 | -0.014 | 0.0716 | No |
| 99 | Tnfrsf9 | 9929 | -0.022 | 0.0611 | No |
| 100 | Hes1 | 10046 | -0.026 | 0.0557 | No |
| 101 | Tnfaip2 | 10283 | -0.035 | 0.0443 | No |
| 102 | Sod2 | 10524 | -0.044 | 0.0329 | No |
| 103 | Ccnd1 | 10616 | -0.048 | 0.0293 | No |
| 104 | Fut4 | 10739 | -0.053 | 0.0242 | No |
| 105 | Irf1 | 10778 | -0.054 | 0.0235 | No |
| 106 | Panx1 | 10848 | -0.057 | 0.0212 | No |
| 107 | Socs3 | 10936 | -0.061 | 0.0181 | No |
| 108 | Atf3 | 11177 | -0.071 | 0.0074 | No |
| 109 | Ptpre | 11200 | -0.072 | 0.0079 | No |
| 110 | Serpinb2 | 11295 | -0.076 | 0.0048 | No |
| 111 | Rel | 11546 | -0.084 | -0.0062 | No |
| 112 | Pde4b | 11629 | -0.087 | -0.0084 | No |
| 113 | Ldlr | 11826 | -0.094 | -0.0164 | No |
| 114 | Gem | 11854 | -0.096 | -0.0156 | No |
| 115 | Ripk2 | 11944 | -0.099 | -0.0179 | No |
| 116 | Btg3 | 11950 | -0.099 | -0.0159 | No |
| 117 | Il18 | 11968 | -0.100 | -0.0145 | No |
| 118 | Snn | 12005 | -0.101 | -0.0140 | No |
| 119 | G0s2 | 12167 | -0.108 | -0.0199 | No |
| 120 | Lamb3 | 12216 | -0.110 | -0.0198 | No |
| 121 | Fosl1 | 12319 | -0.115 | -0.0224 | No |
| 122 | Ccl5 | 12451 | -0.121 | -0.0264 | No |
| 123 | Icam1 | 12588 | -0.125 | -0.0306 | No |
| 124 | Csf1 | 12827 | -0.137 | -0.0397 | No |
| 125 | Eif1 | 13293 | -0.156 | -0.0602 | No |
| 126 | Cdkn1a | 13574 | -0.170 | -0.0708 | No |
| 127 | Nfat5 | 13651 | -0.174 | -0.0707 | No |
| 128 | Bcl3 | 13687 | -0.176 | -0.0684 | No |
| 129 | Cd83 | 13713 | -0.177 | -0.0657 | No |
| 130 | Rela | 13978 | -0.189 | -0.0750 | No |
| 131 | Rcan1 | 14444 | -0.211 | -0.0942 | No |
| 132 | Cflar | 14599 | -0.218 | -0.0971 | No |
| 133 | Pnrc1 | 14615 | -0.219 | -0.0929 | No |
| 134 | Nfil3 | 14952 | -0.237 | -0.1048 | No |
| 135 | Nfkbia | 14970 | -0.238 | -0.1002 | No |
| 136 | Nfkb1 | 15112 | -0.246 | -0.1018 | No |
| 137 | Tank | 15655 | -0.275 | -0.1235 | No |
| 138 | Ptgs2 | 16086 | -0.301 | -0.1388 | No |
| 139 | Ptger4 | 16268 | -0.314 | -0.1410 | No |
| 140 | Junb | 16284 | -0.314 | -0.1345 | No |
| 141 | Rnf19b | 16379 | -0.321 | -0.1320 | No |
| 142 | Slc2a3 | 16403 | -0.324 | -0.1257 | No |
| 143 | Cebpd | 16413 | -0.324 | -0.1188 | No |
| 144 | Spsb1 | 16611 | -0.337 | -0.1212 | No |
| 145 | Tnf | 16839 | -0.353 | -0.1248 | No |
| 146 | Ifngr2 | 16880 | -0.355 | -0.1187 | No |
| 147 | Sphk1 | 17006 | -0.365 | -0.1168 | No |
| 148 | Plau | 17172 | -0.377 | -0.1166 | No |
| 149 | Tgif1 | 17359 | -0.391 | -0.1173 | No |
| 150 | Litaf | 17518 | -0.407 | -0.1161 | No |
| 151 | Tnfaip3 | 17582 | -0.413 | -0.1098 | No |
| 152 | Relb | 17636 | -0.418 | -0.1030 | No |
| 153 | Gch1 | 17675 | -0.422 | -0.0952 | No |
| 154 | Clcf1 | 17707 | -0.427 | -0.0870 | No |
| 155 | Mxd1 | 17817 | -0.439 | -0.0825 | No |
| 156 | Klf6 | 17936 | -0.451 | -0.0783 | No |
| 157 | Sqstm1 | 17987 | -0.456 | -0.0704 | No |
| 158 | Dusp2 | 18331 | -0.502 | -0.0766 | No |
| 159 | Cd69 | 18357 | -0.506 | -0.0662 | No |
| 160 | Olr1 | 18360 | -0.506 | -0.0547 | No |
| 161 | Ier3 | 18498 | -0.527 | -0.0497 | No |
| 162 | Traf1 | 18628 | -0.551 | -0.0437 | No |
| 163 | Gadd45a | 18739 | -0.572 | -0.0362 | No |
| 164 | Marcks | 18751 | -0.574 | -0.0236 | No |
| 165 | Plk2 | 18801 | -0.582 | -0.0128 | No |
| 166 | Ier5 | 19080 | -0.668 | -0.0118 | No |
| 167 | Ets2 | 19176 | -0.703 | -0.0005 | No |
| 168 | Icosl | 19300 | -0.777 | 0.0110 | No |