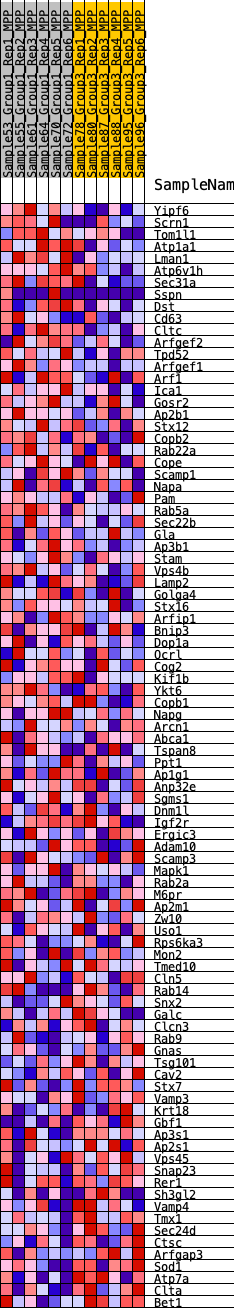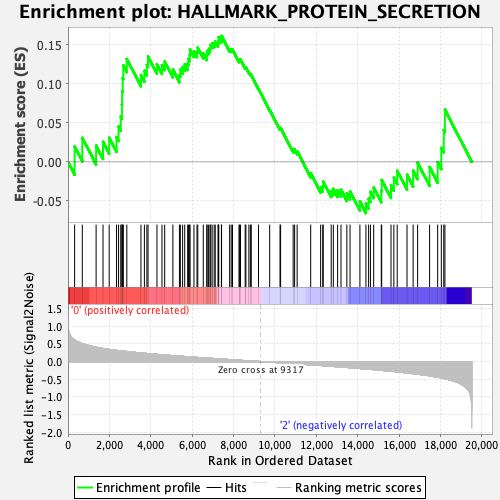
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group3.MPP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
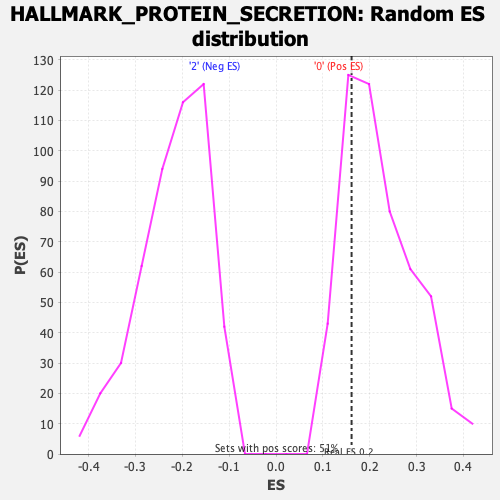
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.16137868 |
| Normalized Enrichment Score (NES) | 0.73298717 |
| Nominal p-value | 0.7519685 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Yipf6 | 320 | 0.609 | 0.0198 | Yes |
| 2 | Scrn1 | 691 | 0.505 | 0.0309 | Yes |
| 3 | Tom1l1 | 1358 | 0.407 | 0.0209 | Yes |
| 4 | Atp1a1 | 1692 | 0.370 | 0.0258 | Yes |
| 5 | Lman1 | 1990 | 0.345 | 0.0311 | Yes |
| 6 | Atp6v1h | 2345 | 0.317 | 0.0318 | Yes |
| 7 | Sec31a | 2444 | 0.311 | 0.0453 | Yes |
| 8 | Sspn | 2549 | 0.305 | 0.0581 | Yes |
| 9 | Dst | 2605 | 0.302 | 0.0733 | Yes |
| 10 | Cd63 | 2616 | 0.302 | 0.0908 | Yes |
| 11 | Cltc | 2643 | 0.299 | 0.1073 | Yes |
| 12 | Arfgef2 | 2672 | 0.298 | 0.1236 | Yes |
| 13 | Tpd52 | 2842 | 0.285 | 0.1319 | Yes |
| 14 | Arfgef1 | 3525 | 0.244 | 0.1113 | Yes |
| 15 | Arf1 | 3691 | 0.235 | 0.1168 | Yes |
| 16 | Ica1 | 3812 | 0.229 | 0.1243 | Yes |
| 17 | Gosr2 | 3867 | 0.227 | 0.1350 | Yes |
| 18 | Ap2b1 | 4298 | 0.204 | 0.1250 | Yes |
| 19 | Stx12 | 4541 | 0.193 | 0.1241 | Yes |
| 20 | Copb2 | 4671 | 0.187 | 0.1286 | Yes |
| 21 | Rab22a | 5068 | 0.168 | 0.1182 | Yes |
| 22 | Cope | 5385 | 0.156 | 0.1112 | Yes |
| 23 | Scamp1 | 5428 | 0.155 | 0.1183 | Yes |
| 24 | Napa | 5538 | 0.149 | 0.1216 | Yes |
| 25 | Pam | 5639 | 0.144 | 0.1250 | Yes |
| 26 | Rab5a | 5784 | 0.137 | 0.1258 | Yes |
| 27 | Sec22b | 5820 | 0.135 | 0.1320 | Yes |
| 28 | Gla | 5883 | 0.132 | 0.1367 | Yes |
| 29 | Ap3b1 | 5894 | 0.132 | 0.1441 | Yes |
| 30 | Stam | 6086 | 0.123 | 0.1416 | Yes |
| 31 | Vps4b | 6233 | 0.118 | 0.1411 | Yes |
| 32 | Lamp2 | 6271 | 0.117 | 0.1461 | Yes |
| 33 | Golga4 | 6536 | 0.109 | 0.1390 | Yes |
| 34 | Stx16 | 6705 | 0.101 | 0.1364 | Yes |
| 35 | Arfip1 | 6721 | 0.101 | 0.1417 | Yes |
| 36 | Bnip3 | 6793 | 0.097 | 0.1438 | Yes |
| 37 | Dop1a | 6861 | 0.094 | 0.1460 | Yes |
| 38 | Ocrl | 6896 | 0.093 | 0.1498 | Yes |
| 39 | Cog2 | 6965 | 0.090 | 0.1517 | Yes |
| 40 | Kif1b | 7063 | 0.086 | 0.1518 | Yes |
| 41 | Ykt6 | 7114 | 0.084 | 0.1543 | Yes |
| 42 | Copb1 | 7249 | 0.079 | 0.1521 | Yes |
| 43 | Napg | 7274 | 0.078 | 0.1555 | Yes |
| 44 | Arcn1 | 7283 | 0.078 | 0.1597 | Yes |
| 45 | Abca1 | 7412 | 0.072 | 0.1575 | Yes |
| 46 | Tspan8 | 7420 | 0.072 | 0.1614 | Yes |
| 47 | Ppt1 | 7811 | 0.056 | 0.1446 | No |
| 48 | Ap1g1 | 7886 | 0.053 | 0.1440 | No |
| 49 | Anp32e | 7949 | 0.051 | 0.1438 | No |
| 50 | Sgms1 | 8267 | 0.040 | 0.1299 | No |
| 51 | Dnm1l | 8280 | 0.039 | 0.1316 | No |
| 52 | Igf2r | 8335 | 0.036 | 0.1310 | No |
| 53 | Ergic3 | 8561 | 0.027 | 0.1210 | No |
| 54 | Adam10 | 8596 | 0.026 | 0.1208 | No |
| 55 | Scamp3 | 8740 | 0.022 | 0.1148 | No |
| 56 | Mapk1 | 8812 | 0.019 | 0.1122 | No |
| 57 | Rab2a | 8855 | 0.017 | 0.1111 | No |
| 58 | M6pr | 9204 | 0.005 | 0.0935 | No |
| 59 | Ap2m1 | 9745 | -0.015 | 0.0665 | No |
| 60 | Zw10 | 10245 | -0.034 | 0.0429 | No |
| 61 | Uso1 | 10273 | -0.035 | 0.0436 | No |
| 62 | Rps6ka3 | 10880 | -0.059 | 0.0159 | No |
| 63 | Mon2 | 10950 | -0.062 | 0.0160 | No |
| 64 | Tmed10 | 11072 | -0.067 | 0.0138 | No |
| 65 | Cln5 | 11727 | -0.091 | -0.0145 | No |
| 66 | Rab14 | 12207 | -0.110 | -0.0326 | No |
| 67 | Snx2 | 12311 | -0.115 | -0.0311 | No |
| 68 | Galc | 12330 | -0.115 | -0.0251 | No |
| 69 | Clcn3 | 12718 | -0.132 | -0.0372 | No |
| 70 | Rab9 | 12821 | -0.137 | -0.0343 | No |
| 71 | Gnas | 13028 | -0.146 | -0.0362 | No |
| 72 | Tsg101 | 13191 | -0.152 | -0.0355 | No |
| 73 | Cav2 | 13475 | -0.165 | -0.0403 | No |
| 74 | Stx7 | 13628 | -0.173 | -0.0378 | No |
| 75 | Vamp3 | 14105 | -0.196 | -0.0506 | No |
| 76 | Krt18 | 14396 | -0.210 | -0.0531 | No |
| 77 | Gbf1 | 14525 | -0.215 | -0.0468 | No |
| 78 | Ap3s1 | 14614 | -0.219 | -0.0383 | No |
| 79 | Ap2s1 | 14771 | -0.227 | -0.0328 | No |
| 80 | Vps45 | 15137 | -0.247 | -0.0369 | No |
| 81 | Snap23 | 15158 | -0.249 | -0.0231 | No |
| 82 | Rer1 | 15607 | -0.273 | -0.0298 | No |
| 83 | Sh3gl2 | 15745 | -0.280 | -0.0202 | No |
| 84 | Vamp4 | 15907 | -0.290 | -0.0112 | No |
| 85 | Tmx1 | 16381 | -0.322 | -0.0163 | No |
| 86 | Sec24d | 16676 | -0.343 | -0.0111 | No |
| 87 | Ctsc | 16893 | -0.356 | -0.0009 | No |
| 88 | Arfgap3 | 17472 | -0.403 | -0.0067 | No |
| 89 | Sod1 | 17862 | -0.444 | -0.0002 | No |
| 90 | Atp7a | 18039 | -0.461 | 0.0182 | No |
| 91 | Clta | 18157 | -0.480 | 0.0407 | No |
| 92 | Bet1 | 18212 | -0.486 | 0.0669 | No |