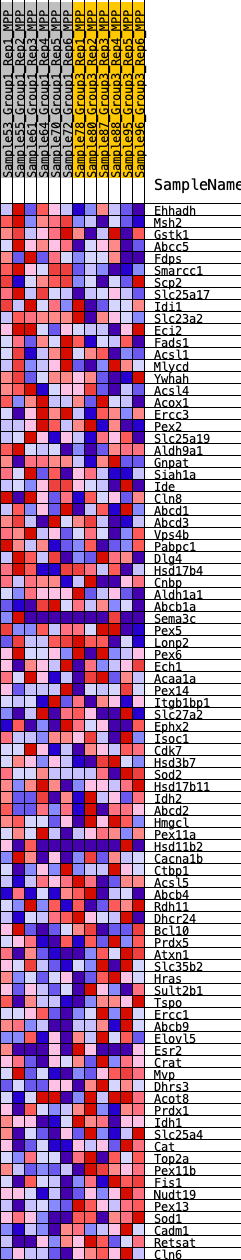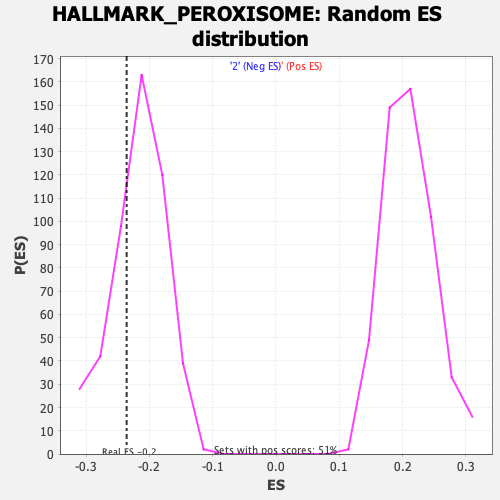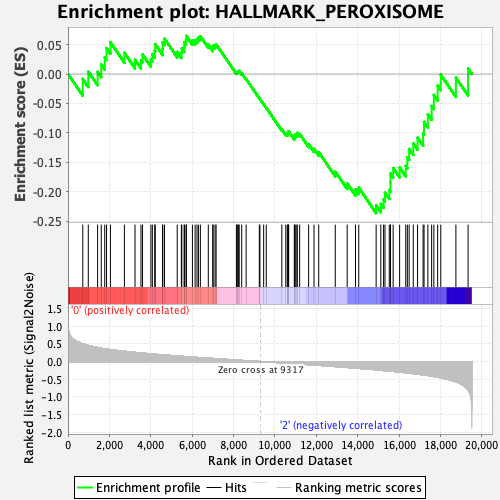
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group3.MPP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.2360928 |
| Normalized Enrichment Score (NES) | -1.0891823 |
| Nominal p-value | 0.2906504 |
| FDR q-value | 0.9684608 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ehhadh | 714 | 0.500 | -0.0083 | No |
| 2 | Msh2 | 980 | 0.458 | 0.0041 | No |
| 3 | Gstk1 | 1431 | 0.398 | 0.0036 | No |
| 4 | Abcc5 | 1607 | 0.379 | 0.0162 | No |
| 5 | Fdps | 1775 | 0.363 | 0.0283 | No |
| 6 | Smarcc1 | 1863 | 0.357 | 0.0441 | No |
| 7 | Scp2 | 2050 | 0.339 | 0.0539 | No |
| 8 | Slc25a17 | 2725 | 0.294 | 0.0359 | No |
| 9 | Idi1 | 3238 | 0.259 | 0.0243 | No |
| 10 | Slc23a2 | 3526 | 0.244 | 0.0234 | No |
| 11 | Eci2 | 3601 | 0.240 | 0.0332 | No |
| 12 | Fads1 | 4006 | 0.220 | 0.0249 | No |
| 13 | Acsl1 | 4076 | 0.216 | 0.0337 | No |
| 14 | Mlycd | 4191 | 0.209 | 0.0397 | No |
| 15 | Ywhah | 4215 | 0.208 | 0.0504 | No |
| 16 | Acsl4 | 4573 | 0.191 | 0.0429 | No |
| 17 | Acox1 | 4576 | 0.191 | 0.0537 | No |
| 18 | Ercc3 | 4661 | 0.187 | 0.0600 | No |
| 19 | Pex2 | 5277 | 0.161 | 0.0375 | No |
| 20 | Slc25a19 | 5478 | 0.152 | 0.0358 | No |
| 21 | Aldh9a1 | 5499 | 0.151 | 0.0434 | No |
| 22 | Gnpat | 5616 | 0.145 | 0.0457 | No |
| 23 | Siah1a | 5621 | 0.145 | 0.0538 | No |
| 24 | Ide | 5703 | 0.141 | 0.0576 | No |
| 25 | Cln8 | 5720 | 0.140 | 0.0647 | No |
| 26 | Abcd1 | 6012 | 0.126 | 0.0570 | No |
| 27 | Abcd3 | 6142 | 0.121 | 0.0572 | No |
| 28 | Vps4b | 6233 | 0.118 | 0.0593 | No |
| 29 | Pabpc1 | 6303 | 0.116 | 0.0624 | No |
| 30 | Dlg4 | 6396 | 0.114 | 0.0641 | No |
| 31 | Hsd17b4 | 6779 | 0.098 | 0.0500 | No |
| 32 | Cnbp | 6996 | 0.089 | 0.0440 | No |
| 33 | Aldh1a1 | 7012 | 0.088 | 0.0482 | No |
| 34 | Abcb1a | 7104 | 0.085 | 0.0484 | No |
| 35 | Sema3c | 7156 | 0.083 | 0.0505 | No |
| 36 | Pex5 | 8137 | 0.045 | 0.0026 | No |
| 37 | Lonp2 | 8167 | 0.043 | 0.0035 | No |
| 38 | Pex6 | 8206 | 0.042 | 0.0040 | No |
| 39 | Ech1 | 8247 | 0.040 | 0.0042 | No |
| 40 | Acaa1a | 8268 | 0.039 | 0.0054 | No |
| 41 | Pex14 | 8393 | 0.034 | 0.0010 | No |
| 42 | Itgb1bp1 | 8607 | 0.026 | -0.0085 | No |
| 43 | Slc27a2 | 9244 | 0.003 | -0.0410 | No |
| 44 | Ephx2 | 9274 | 0.002 | -0.0424 | No |
| 45 | Isoc1 | 9454 | -0.004 | -0.0514 | No |
| 46 | Cdk7 | 9580 | -0.008 | -0.0574 | No |
| 47 | Hsd3b7 | 10332 | -0.037 | -0.0939 | No |
| 48 | Sod2 | 10524 | -0.044 | -0.1012 | No |
| 49 | Hsd17b11 | 10598 | -0.047 | -0.1023 | No |
| 50 | Idh2 | 10624 | -0.049 | -0.1008 | No |
| 51 | Abcd2 | 10655 | -0.050 | -0.0995 | No |
| 52 | Hmgcl | 10663 | -0.050 | -0.0971 | No |
| 53 | Pex11a | 10935 | -0.061 | -0.1075 | No |
| 54 | Hsd11b2 | 10952 | -0.062 | -0.1048 | No |
| 55 | Cacna1b | 10981 | -0.063 | -0.1027 | No |
| 56 | Ctbp1 | 11061 | -0.067 | -0.1030 | No |
| 57 | Acsl5 | 11074 | -0.067 | -0.0998 | No |
| 58 | Abcb4 | 11194 | -0.072 | -0.1018 | No |
| 59 | Rdh11 | 11627 | -0.087 | -0.1191 | No |
| 60 | Dhcr24 | 11887 | -0.097 | -0.1269 | No |
| 61 | Bcl10 | 12115 | -0.106 | -0.1325 | No |
| 62 | Prdx5 | 12914 | -0.141 | -0.1656 | No |
| 63 | Atxn1 | 13490 | -0.166 | -0.1857 | No |
| 64 | Slc35b2 | 13892 | -0.185 | -0.1958 | No |
| 65 | Hras | 14049 | -0.193 | -0.1929 | No |
| 66 | Sult2b1 | 14889 | -0.233 | -0.2228 | Yes |
| 67 | Tspo | 15116 | -0.247 | -0.2204 | Yes |
| 68 | Ercc1 | 15249 | -0.255 | -0.2127 | Yes |
| 69 | Abcb9 | 15316 | -0.258 | -0.2014 | Yes |
| 70 | Elovl5 | 15524 | -0.268 | -0.1968 | Yes |
| 71 | Esr2 | 15580 | -0.272 | -0.1842 | Yes |
| 72 | Crat | 15583 | -0.272 | -0.1688 | Yes |
| 73 | Mvp | 15711 | -0.278 | -0.1595 | Yes |
| 74 | Dhrs3 | 16024 | -0.297 | -0.1586 | Yes |
| 75 | Acot8 | 16320 | -0.316 | -0.1558 | Yes |
| 76 | Prdx1 | 16397 | -0.323 | -0.1414 | Yes |
| 77 | Idh1 | 16484 | -0.327 | -0.1271 | Yes |
| 78 | Slc25a4 | 16683 | -0.343 | -0.1178 | Yes |
| 79 | Cat | 16890 | -0.356 | -0.1081 | Yes |
| 80 | Top2a | 17158 | -0.376 | -0.1005 | Yes |
| 81 | Pex11b | 17204 | -0.379 | -0.0813 | Yes |
| 82 | Fis1 | 17389 | -0.394 | -0.0683 | Yes |
| 83 | Nudt19 | 17571 | -0.412 | -0.0542 | Yes |
| 84 | Pex13 | 17678 | -0.423 | -0.0356 | Yes |
| 85 | Sod1 | 17862 | -0.444 | -0.0197 | Yes |
| 86 | Cadm1 | 18012 | -0.459 | -0.0012 | Yes |
| 87 | Retsat | 18743 | -0.573 | -0.0062 | Yes |
| 88 | Cln6 | 19333 | -0.803 | 0.0092 | Yes |