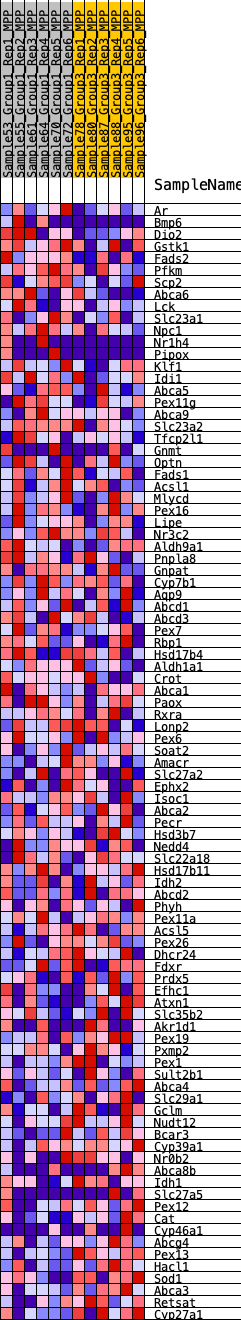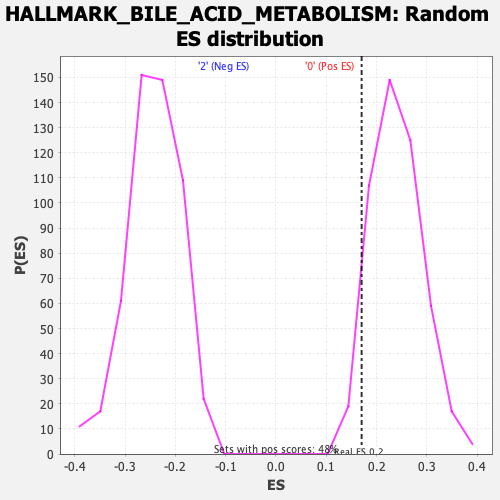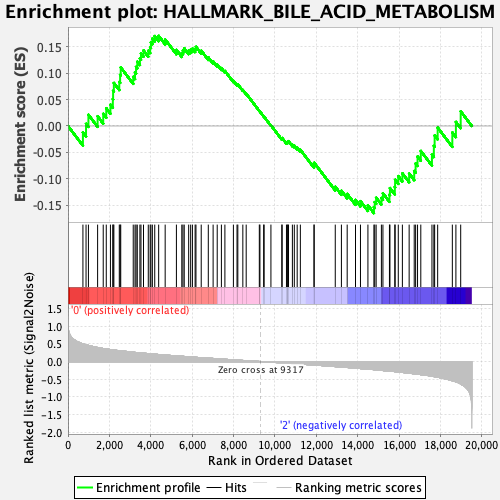
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group3.MPP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.17033465 |
| Normalized Enrichment Score (NES) | 0.7060644 |
| Nominal p-value | 0.94166666 |
| FDR q-value | 0.963816 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ar | 721 | 0.499 | -0.0120 | Yes |
| 2 | Bmp6 | 872 | 0.476 | 0.0041 | Yes |
| 3 | Dio2 | 987 | 0.457 | 0.0212 | Yes |
| 4 | Gstk1 | 1431 | 0.398 | 0.0184 | Yes |
| 5 | Fads2 | 1702 | 0.369 | 0.0231 | Yes |
| 6 | Pfkm | 1849 | 0.358 | 0.0335 | Yes |
| 7 | Scp2 | 2050 | 0.339 | 0.0403 | Yes |
| 8 | Abca6 | 2167 | 0.330 | 0.0509 | Yes |
| 9 | Lck | 2180 | 0.328 | 0.0668 | Yes |
| 10 | Slc23a1 | 2216 | 0.325 | 0.0813 | Yes |
| 11 | Npc1 | 2478 | 0.309 | 0.0834 | Yes |
| 12 | Nr1h4 | 2519 | 0.306 | 0.0968 | Yes |
| 13 | Pipox | 2548 | 0.305 | 0.1107 | Yes |
| 14 | Klf1 | 3152 | 0.265 | 0.0929 | Yes |
| 15 | Idi1 | 3238 | 0.259 | 0.1016 | Yes |
| 16 | Abca5 | 3289 | 0.256 | 0.1119 | Yes |
| 17 | Pex11g | 3348 | 0.252 | 0.1216 | Yes |
| 18 | Abca9 | 3474 | 0.246 | 0.1275 | Yes |
| 19 | Slc23a2 | 3526 | 0.244 | 0.1371 | Yes |
| 20 | Tfcp2l1 | 3648 | 0.236 | 0.1428 | Yes |
| 21 | Gnmt | 3879 | 0.226 | 0.1423 | Yes |
| 22 | Optn | 3970 | 0.221 | 0.1488 | Yes |
| 23 | Fads1 | 4006 | 0.220 | 0.1580 | Yes |
| 24 | Acsl1 | 4076 | 0.216 | 0.1653 | Yes |
| 25 | Mlycd | 4191 | 0.209 | 0.1700 | Yes |
| 26 | Pex16 | 4381 | 0.200 | 0.1703 | Yes |
| 27 | Lipe | 4695 | 0.185 | 0.1635 | No |
| 28 | Nr3c2 | 5238 | 0.163 | 0.1438 | No |
| 29 | Aldh9a1 | 5499 | 0.151 | 0.1380 | No |
| 30 | Pnpla8 | 5554 | 0.148 | 0.1427 | No |
| 31 | Gnpat | 5616 | 0.145 | 0.1469 | No |
| 32 | Cyp7b1 | 5831 | 0.135 | 0.1426 | No |
| 33 | Aqp9 | 5927 | 0.130 | 0.1443 | No |
| 34 | Abcd1 | 6012 | 0.126 | 0.1463 | No |
| 35 | Abcd3 | 6142 | 0.121 | 0.1458 | No |
| 36 | Pex7 | 6183 | 0.120 | 0.1497 | No |
| 37 | Rbp1 | 6437 | 0.112 | 0.1424 | No |
| 38 | Hsd17b4 | 6779 | 0.098 | 0.1297 | No |
| 39 | Aldh1a1 | 7012 | 0.088 | 0.1222 | No |
| 40 | Crot | 7210 | 0.081 | 0.1162 | No |
| 41 | Abca1 | 7412 | 0.072 | 0.1094 | No |
| 42 | Paox | 7581 | 0.065 | 0.1040 | No |
| 43 | Rxra | 7992 | 0.049 | 0.0854 | No |
| 44 | Lonp2 | 8167 | 0.043 | 0.0786 | No |
| 45 | Pex6 | 8206 | 0.042 | 0.0788 | No |
| 46 | Soat2 | 8448 | 0.032 | 0.0680 | No |
| 47 | Amacr | 8610 | 0.026 | 0.0610 | No |
| 48 | Slc27a2 | 9244 | 0.003 | 0.0286 | No |
| 49 | Ephx2 | 9274 | 0.002 | 0.0272 | No |
| 50 | Isoc1 | 9454 | -0.004 | 0.0182 | No |
| 51 | Abca2 | 9476 | -0.005 | 0.0174 | No |
| 52 | Pecr | 9805 | -0.017 | 0.0013 | No |
| 53 | Hsd3b7 | 10332 | -0.037 | -0.0239 | No |
| 54 | Nedd4 | 10346 | -0.038 | -0.0227 | No |
| 55 | Slc22a18 | 10556 | -0.046 | -0.0311 | No |
| 56 | Hsd17b11 | 10598 | -0.047 | -0.0308 | No |
| 57 | Idh2 | 10624 | -0.049 | -0.0297 | No |
| 58 | Abcd2 | 10655 | -0.050 | -0.0287 | No |
| 59 | Phyh | 10842 | -0.057 | -0.0354 | No |
| 60 | Pex11a | 10935 | -0.061 | -0.0371 | No |
| 61 | Acsl5 | 11074 | -0.067 | -0.0409 | No |
| 62 | Pex26 | 11227 | -0.074 | -0.0450 | No |
| 63 | Dhcr24 | 11887 | -0.097 | -0.0740 | No |
| 64 | Fdxr | 11894 | -0.097 | -0.0694 | No |
| 65 | Prdx5 | 12914 | -0.141 | -0.1148 | No |
| 66 | Efhc1 | 13211 | -0.153 | -0.1224 | No |
| 67 | Atxn1 | 13490 | -0.166 | -0.1284 | No |
| 68 | Slc35b2 | 13892 | -0.185 | -0.1397 | No |
| 69 | Akr1d1 | 14132 | -0.197 | -0.1421 | No |
| 70 | Pex19 | 14492 | -0.213 | -0.1499 | No |
| 71 | Pxmp2 | 14781 | -0.227 | -0.1533 | No |
| 72 | Pex1 | 14822 | -0.229 | -0.1438 | No |
| 73 | Sult2b1 | 14889 | -0.233 | -0.1355 | No |
| 74 | Abca4 | 15143 | -0.248 | -0.1361 | No |
| 75 | Slc29a1 | 15217 | -0.252 | -0.1271 | No |
| 76 | Gclm | 15532 | -0.269 | -0.1297 | No |
| 77 | Nudt12 | 15556 | -0.270 | -0.1173 | No |
| 78 | Bcar3 | 15792 | -0.283 | -0.1152 | No |
| 79 | Cyp39a1 | 15806 | -0.284 | -0.1016 | No |
| 80 | Nr0b2 | 15959 | -0.293 | -0.0947 | No |
| 81 | Abca8b | 16157 | -0.305 | -0.0895 | No |
| 82 | Idh1 | 16484 | -0.327 | -0.0899 | No |
| 83 | Slc27a5 | 16729 | -0.347 | -0.0850 | No |
| 84 | Pex12 | 16797 | -0.351 | -0.0708 | No |
| 85 | Cat | 16890 | -0.356 | -0.0576 | No |
| 86 | Cyp46a1 | 17047 | -0.368 | -0.0472 | No |
| 87 | Abcg4 | 17586 | -0.414 | -0.0541 | No |
| 88 | Pex13 | 17678 | -0.423 | -0.0375 | No |
| 89 | Hacl1 | 17716 | -0.427 | -0.0179 | No |
| 90 | Sod1 | 17862 | -0.444 | -0.0030 | No |
| 91 | Abca3 | 18571 | -0.542 | -0.0123 | No |
| 92 | Retsat | 18743 | -0.573 | 0.0077 | No |
| 93 | Cyp27a1 | 18975 | -0.633 | 0.0277 | No |