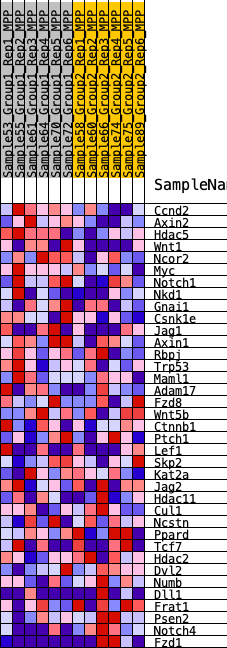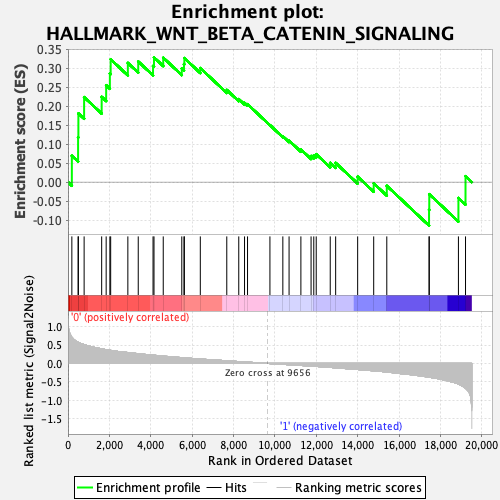
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.32844013 |
| Normalized Enrichment Score (NES) | 1.1601851 |
| Nominal p-value | 0.24710424 |
| FDR q-value | 0.89303994 |
| FWER p-Value | 0.953 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ccnd2 | 186 | 0.710 | 0.0695 | Yes |
| 2 | Axin2 | 488 | 0.575 | 0.1181 | Yes |
| 3 | Hdac5 | 502 | 0.571 | 0.1810 | Yes |
| 4 | Wnt1 | 779 | 0.512 | 0.2239 | Yes |
| 5 | Ncor2 | 1627 | 0.397 | 0.2245 | Yes |
| 6 | Myc | 1844 | 0.374 | 0.2551 | Yes |
| 7 | Notch1 | 2023 | 0.359 | 0.2859 | Yes |
| 8 | Nkd1 | 2062 | 0.355 | 0.3235 | Yes |
| 9 | Gnai1 | 2892 | 0.297 | 0.3140 | Yes |
| 10 | Csnk1e | 3394 | 0.264 | 0.3177 | Yes |
| 11 | Jag1 | 4109 | 0.224 | 0.3060 | Yes |
| 12 | Axin1 | 4155 | 0.222 | 0.3284 | Yes |
| 13 | Rbpj | 4602 | 0.199 | 0.3277 | No |
| 14 | Trp53 | 5498 | 0.157 | 0.2993 | No |
| 15 | Maml1 | 5604 | 0.152 | 0.3108 | No |
| 16 | Adam17 | 5623 | 0.151 | 0.3267 | No |
| 17 | Fzd8 | 6393 | 0.118 | 0.3004 | No |
| 18 | Wnt5b | 7672 | 0.072 | 0.2428 | No |
| 19 | Ctnnb1 | 8251 | 0.050 | 0.2186 | No |
| 20 | Ptch1 | 8530 | 0.040 | 0.2088 | No |
| 21 | Lef1 | 8675 | 0.035 | 0.2053 | No |
| 22 | Skp2 | 9757 | -0.003 | 0.1501 | No |
| 23 | Kat2a | 10381 | -0.024 | 0.1208 | No |
| 24 | Jag2 | 10682 | -0.034 | 0.1093 | No |
| 25 | Hdac11 | 11253 | -0.054 | 0.0860 | No |
| 26 | Cul1 | 11745 | -0.072 | 0.0688 | No |
| 27 | Ncstn | 11876 | -0.076 | 0.0706 | No |
| 28 | Ppard | 11994 | -0.081 | 0.0736 | No |
| 29 | Tcf7 | 12671 | -0.107 | 0.0508 | No |
| 30 | Hdac2 | 12933 | -0.119 | 0.0506 | No |
| 31 | Dvl2 | 13996 | -0.164 | 0.0144 | No |
| 32 | Numb | 14771 | -0.200 | -0.0031 | No |
| 33 | Dll1 | 15404 | -0.232 | -0.0097 | No |
| 34 | Frat1 | 17446 | -0.373 | -0.0729 | No |
| 35 | Psen2 | 17457 | -0.374 | -0.0318 | No |
| 36 | Notch4 | 18865 | -0.559 | -0.0418 | No |
| 37 | Fzd1 | 19208 | -0.673 | 0.0156 | No |