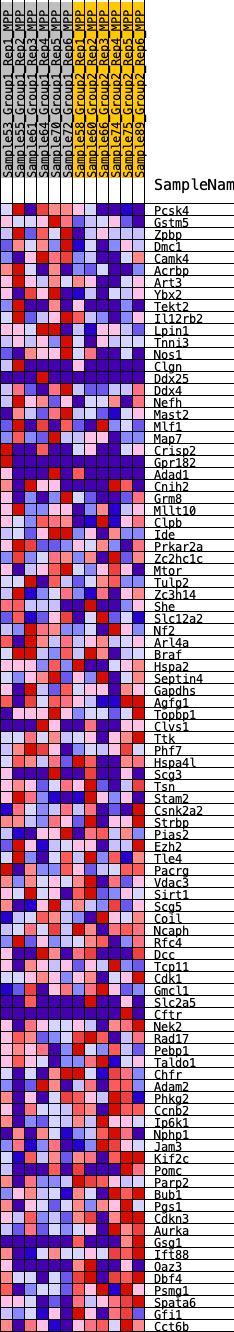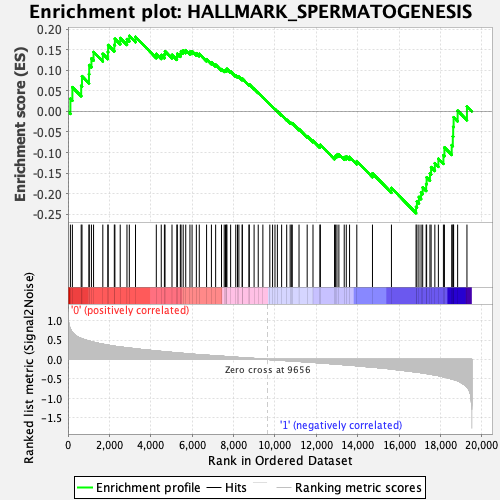
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.24728319 |
| Normalized Enrichment Score (NES) | -1.0254289 |
| Nominal p-value | 0.448 |
| FDR q-value | 0.83713907 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pcsk4 | 116 | 0.780 | 0.0309 | No |
| 2 | Gstm5 | 212 | 0.694 | 0.0588 | No |
| 3 | Zpbp | 637 | 0.539 | 0.0624 | No |
| 4 | Dmc1 | 676 | 0.532 | 0.0856 | No |
| 5 | Camk4 | 1014 | 0.475 | 0.0907 | No |
| 6 | Acrbp | 1024 | 0.474 | 0.1126 | No |
| 7 | Art3 | 1132 | 0.457 | 0.1287 | No |
| 8 | Ybx2 | 1235 | 0.441 | 0.1443 | No |
| 9 | Tekt2 | 1682 | 0.391 | 0.1398 | No |
| 10 | Il12rb2 | 1927 | 0.367 | 0.1445 | No |
| 11 | Lpin1 | 1939 | 0.365 | 0.1612 | No |
| 12 | Tnni3 | 2238 | 0.339 | 0.1619 | No |
| 13 | Nos1 | 2266 | 0.337 | 0.1764 | No |
| 14 | Clgn | 2523 | 0.317 | 0.1782 | No |
| 15 | Ddx25 | 2849 | 0.299 | 0.1756 | No |
| 16 | Ddx4 | 2963 | 0.291 | 0.1836 | No |
| 17 | Nefh | 3262 | 0.273 | 0.1811 | No |
| 18 | Mast2 | 4267 | 0.217 | 0.1397 | No |
| 19 | Mlf1 | 4506 | 0.205 | 0.1371 | No |
| 20 | Map7 | 4658 | 0.196 | 0.1386 | No |
| 21 | Crisp2 | 4688 | 0.194 | 0.1462 | No |
| 22 | Gpr182 | 5025 | 0.179 | 0.1374 | No |
| 23 | Adad1 | 5261 | 0.168 | 0.1333 | No |
| 24 | Cnih2 | 5281 | 0.168 | 0.1402 | No |
| 25 | Grm8 | 5428 | 0.160 | 0.1402 | No |
| 26 | Mllt10 | 5462 | 0.159 | 0.1460 | No |
| 27 | Clpb | 5566 | 0.154 | 0.1480 | No |
| 28 | Ide | 5689 | 0.148 | 0.1487 | No |
| 29 | Prkar2a | 5892 | 0.140 | 0.1449 | No |
| 30 | Zc2hc1c | 5995 | 0.135 | 0.1460 | No |
| 31 | Mtor | 6202 | 0.126 | 0.1414 | No |
| 32 | Tulp2 | 6343 | 0.120 | 0.1398 | No |
| 33 | Zc3h14 | 6696 | 0.107 | 0.1268 | No |
| 34 | She | 6930 | 0.099 | 0.1194 | No |
| 35 | Slc12a2 | 7134 | 0.092 | 0.1133 | No |
| 36 | Nf2 | 7417 | 0.082 | 0.1027 | No |
| 37 | Arl4a | 7548 | 0.076 | 0.0996 | No |
| 38 | Braf | 7610 | 0.074 | 0.0999 | No |
| 39 | Hspa2 | 7666 | 0.072 | 0.1005 | No |
| 40 | Septin4 | 7675 | 0.072 | 0.1034 | No |
| 41 | Gapdhs | 7858 | 0.065 | 0.0971 | No |
| 42 | Agfg1 | 8106 | 0.056 | 0.0870 | No |
| 43 | Topbp1 | 8190 | 0.052 | 0.0852 | No |
| 44 | Clvs1 | 8257 | 0.049 | 0.0841 | No |
| 45 | Ttk | 8417 | 0.043 | 0.0780 | No |
| 46 | Phf7 | 8423 | 0.043 | 0.0797 | No |
| 47 | Hspa4l | 8744 | 0.033 | 0.0648 | No |
| 48 | Scg3 | 8758 | 0.033 | 0.0657 | No |
| 49 | Tsn | 8992 | 0.025 | 0.0548 | No |
| 50 | Stam2 | 9192 | 0.017 | 0.0454 | No |
| 51 | Csnk2a2 | 9416 | 0.009 | 0.0343 | No |
| 52 | Strbp | 9752 | -0.003 | 0.0172 | No |
| 53 | Pias2 | 9885 | -0.007 | 0.0108 | No |
| 54 | Ezh2 | 10001 | -0.011 | 0.0054 | No |
| 55 | Tle4 | 10117 | -0.015 | 0.0002 | No |
| 56 | Pacrg | 10322 | -0.022 | -0.0093 | No |
| 57 | Vdac3 | 10567 | -0.030 | -0.0204 | No |
| 58 | Sirt1 | 10734 | -0.036 | -0.0273 | No |
| 59 | Scg5 | 10791 | -0.038 | -0.0283 | No |
| 60 | Coil | 10837 | -0.040 | -0.0288 | No |
| 61 | Ncaph | 11163 | -0.051 | -0.0431 | No |
| 62 | Rfc4 | 11557 | -0.064 | -0.0603 | No |
| 63 | Dcc | 11838 | -0.075 | -0.0712 | No |
| 64 | Tcp11 | 12171 | -0.087 | -0.0842 | No |
| 65 | Cdk1 | 12194 | -0.089 | -0.0811 | No |
| 66 | Gmcl1 | 12879 | -0.116 | -0.1109 | No |
| 67 | Slc2a5 | 12931 | -0.118 | -0.1079 | No |
| 68 | Cftr | 12996 | -0.122 | -0.1054 | No |
| 69 | Nek2 | 13089 | -0.125 | -0.1043 | No |
| 70 | Rad17 | 13347 | -0.136 | -0.1111 | No |
| 71 | Pebp1 | 13447 | -0.141 | -0.1096 | No |
| 72 | Taldo1 | 13605 | -0.147 | -0.1107 | No |
| 73 | Chfr | 13955 | -0.162 | -0.1210 | No |
| 74 | Adam2 | 14712 | -0.198 | -0.1506 | No |
| 75 | Phkg2 | 15629 | -0.247 | -0.1861 | No |
| 76 | Ccnb2 | 16818 | -0.322 | -0.2321 | Yes |
| 77 | Ip6k1 | 16860 | -0.325 | -0.2188 | Yes |
| 78 | Nphp1 | 16956 | -0.333 | -0.2080 | Yes |
| 79 | Jam3 | 17060 | -0.340 | -0.1972 | Yes |
| 80 | Kif2c | 17138 | -0.346 | -0.1848 | Yes |
| 81 | Pomc | 17306 | -0.361 | -0.1763 | Yes |
| 82 | Parp2 | 17329 | -0.363 | -0.1604 | Yes |
| 83 | Bub1 | 17489 | -0.377 | -0.1507 | Yes |
| 84 | Pgs1 | 17545 | -0.383 | -0.1355 | Yes |
| 85 | Cdkn3 | 17726 | -0.398 | -0.1259 | Yes |
| 86 | Aurka | 17897 | -0.416 | -0.1151 | Yes |
| 87 | Gsg1 | 18140 | -0.446 | -0.1065 | Yes |
| 88 | Ift88 | 18189 | -0.450 | -0.0877 | Yes |
| 89 | Oaz3 | 18541 | -0.500 | -0.0821 | Yes |
| 90 | Dbf4 | 18594 | -0.508 | -0.0608 | Yes |
| 91 | Psmg1 | 18612 | -0.511 | -0.0375 | Yes |
| 92 | Spata6 | 18633 | -0.513 | -0.0143 | Yes |
| 93 | Gfi1 | 18827 | -0.548 | 0.0017 | Yes |
| 94 | Cct6b | 19277 | -0.710 | 0.0121 | Yes |