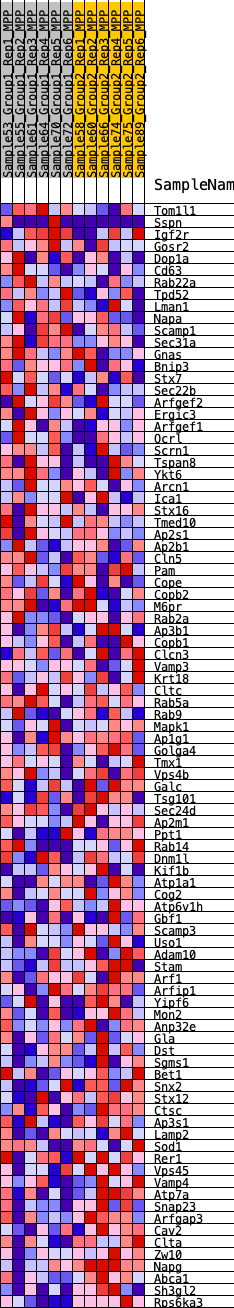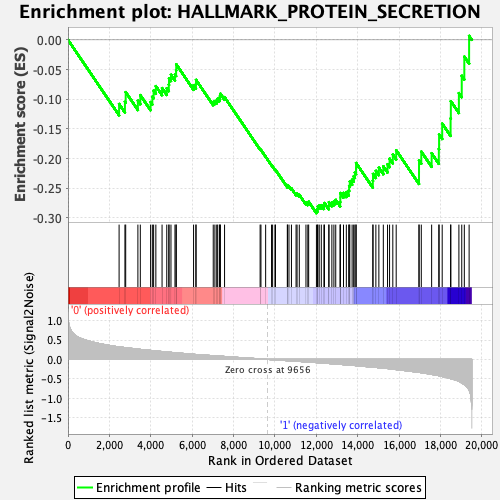
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.29181907 |
| Normalized Enrichment Score (NES) | -1.3727481 |
| Nominal p-value | 0.13387424 |
| FDR q-value | 0.45662245 |
| FWER p-Value | 0.668 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tom1l1 | 2469 | 0.321 | -0.1080 | No |
| 2 | Sspn | 2751 | 0.305 | -0.1042 | No |
| 3 | Igf2r | 2788 | 0.303 | -0.0880 | No |
| 4 | Gosr2 | 3377 | 0.265 | -0.1025 | No |
| 5 | Dop1a | 3494 | 0.258 | -0.0931 | No |
| 6 | Cd63 | 3991 | 0.232 | -0.1048 | No |
| 7 | Rab22a | 4075 | 0.226 | -0.0956 | No |
| 8 | Tpd52 | 4137 | 0.223 | -0.0854 | No |
| 9 | Lman1 | 4245 | 0.218 | -0.0779 | No |
| 10 | Napa | 4545 | 0.202 | -0.0813 | No |
| 11 | Scamp1 | 4772 | 0.191 | -0.0815 | No |
| 12 | Sec31a | 4871 | 0.187 | -0.0754 | No |
| 13 | Gnas | 4882 | 0.186 | -0.0648 | No |
| 14 | Bnip3 | 4975 | 0.181 | -0.0588 | No |
| 15 | Stx7 | 5169 | 0.173 | -0.0584 | No |
| 16 | Sec22b | 5223 | 0.170 | -0.0510 | No |
| 17 | Arfgef2 | 5230 | 0.169 | -0.0412 | No |
| 18 | Ergic3 | 6067 | 0.131 | -0.0764 | No |
| 19 | Arfgef1 | 6181 | 0.126 | -0.0747 | No |
| 20 | Ocrl | 6186 | 0.126 | -0.0674 | No |
| 21 | Scrn1 | 7018 | 0.096 | -0.1044 | No |
| 22 | Tspan8 | 7098 | 0.093 | -0.1029 | No |
| 23 | Ykt6 | 7186 | 0.090 | -0.1020 | No |
| 24 | Arcn1 | 7232 | 0.088 | -0.0991 | No |
| 25 | Ica1 | 7317 | 0.085 | -0.0984 | No |
| 26 | Stx16 | 7344 | 0.084 | -0.0947 | No |
| 27 | Tmed10 | 7364 | 0.083 | -0.0907 | No |
| 28 | Ap2s1 | 7565 | 0.076 | -0.0965 | No |
| 29 | Ap2b1 | 9283 | 0.013 | -0.1841 | No |
| 30 | Cln5 | 9323 | 0.012 | -0.1854 | No |
| 31 | Pam | 9550 | 0.004 | -0.1968 | No |
| 32 | Cope | 9835 | -0.006 | -0.2111 | No |
| 33 | Copb2 | 9887 | -0.007 | -0.2133 | No |
| 34 | M6pr | 9999 | -0.011 | -0.2183 | No |
| 35 | Rab2a | 10026 | -0.012 | -0.2189 | No |
| 36 | Ap3b1 | 10601 | -0.032 | -0.2466 | No |
| 37 | Copb1 | 10602 | -0.032 | -0.2447 | No |
| 38 | Clcn3 | 10677 | -0.034 | -0.2464 | No |
| 39 | Vamp3 | 10797 | -0.038 | -0.2503 | No |
| 40 | Krt18 | 11020 | -0.046 | -0.2590 | No |
| 41 | Cltc | 11068 | -0.048 | -0.2585 | No |
| 42 | Rab5a | 11176 | -0.051 | -0.2610 | No |
| 43 | Rab9 | 11498 | -0.062 | -0.2738 | No |
| 44 | Mapk1 | 11588 | -0.065 | -0.2745 | No |
| 45 | Ap1g1 | 11629 | -0.067 | -0.2726 | No |
| 46 | Golga4 | 12004 | -0.081 | -0.2870 | Yes |
| 47 | Tmx1 | 12055 | -0.083 | -0.2846 | Yes |
| 48 | Vps4b | 12072 | -0.083 | -0.2805 | Yes |
| 49 | Galc | 12139 | -0.086 | -0.2788 | Yes |
| 50 | Tsg101 | 12239 | -0.090 | -0.2785 | Yes |
| 51 | Sec24d | 12366 | -0.096 | -0.2792 | Yes |
| 52 | Ap2m1 | 12393 | -0.097 | -0.2748 | Yes |
| 53 | Ppt1 | 12603 | -0.105 | -0.2793 | Yes |
| 54 | Rab14 | 12609 | -0.105 | -0.2733 | Yes |
| 55 | Dnm1l | 12750 | -0.111 | -0.2739 | Yes |
| 56 | Kif1b | 12848 | -0.115 | -0.2720 | Yes |
| 57 | Atp1a1 | 12937 | -0.119 | -0.2695 | Yes |
| 58 | Cog2 | 13152 | -0.127 | -0.2729 | Yes |
| 59 | Atp6v1h | 13158 | -0.128 | -0.2655 | Yes |
| 60 | Gbf1 | 13159 | -0.128 | -0.2579 | Yes |
| 61 | Scamp3 | 13312 | -0.134 | -0.2578 | Yes |
| 62 | Uso1 | 13449 | -0.141 | -0.2564 | Yes |
| 63 | Adam10 | 13566 | -0.146 | -0.2537 | Yes |
| 64 | Stam | 13586 | -0.146 | -0.2459 | Yes |
| 65 | Arf1 | 13617 | -0.148 | -0.2386 | Yes |
| 66 | Arfip1 | 13728 | -0.151 | -0.2353 | Yes |
| 67 | Yipf6 | 13803 | -0.154 | -0.2299 | Yes |
| 68 | Mon2 | 13862 | -0.157 | -0.2235 | Yes |
| 69 | Anp32e | 13919 | -0.160 | -0.2169 | Yes |
| 70 | Gla | 13920 | -0.160 | -0.2073 | Yes |
| 71 | Dst | 14729 | -0.198 | -0.2371 | Yes |
| 72 | Sgms1 | 14739 | -0.199 | -0.2257 | Yes |
| 73 | Bet1 | 14876 | -0.204 | -0.2206 | Yes |
| 74 | Snx2 | 15021 | -0.213 | -0.2153 | Yes |
| 75 | Stx12 | 15236 | -0.223 | -0.2130 | Yes |
| 76 | Ctsc | 15440 | -0.234 | -0.2095 | Yes |
| 77 | Ap3s1 | 15538 | -0.240 | -0.2002 | Yes |
| 78 | Lamp2 | 15695 | -0.251 | -0.1932 | Yes |
| 79 | Sod1 | 15858 | -0.261 | -0.1860 | Yes |
| 80 | Rer1 | 16961 | -0.334 | -0.2228 | Yes |
| 81 | Vps45 | 16962 | -0.334 | -0.2029 | Yes |
| 82 | Vamp4 | 17071 | -0.341 | -0.1882 | Yes |
| 83 | Atp7a | 17571 | -0.385 | -0.1909 | Yes |
| 84 | Snap23 | 17919 | -0.418 | -0.1838 | Yes |
| 85 | Arfgap3 | 17931 | -0.419 | -0.1594 | Yes |
| 86 | Cav2 | 18081 | -0.438 | -0.1409 | Yes |
| 87 | Clta | 18487 | -0.493 | -0.1323 | Yes |
| 88 | Zw10 | 18499 | -0.495 | -0.1034 | Yes |
| 89 | Napg | 18888 | -0.565 | -0.0896 | Yes |
| 90 | Abca1 | 19026 | -0.606 | -0.0605 | Yes |
| 91 | Sh3gl2 | 19150 | -0.648 | -0.0282 | Yes |
| 92 | Rps6ka3 | 19382 | -0.785 | 0.0067 | Yes |