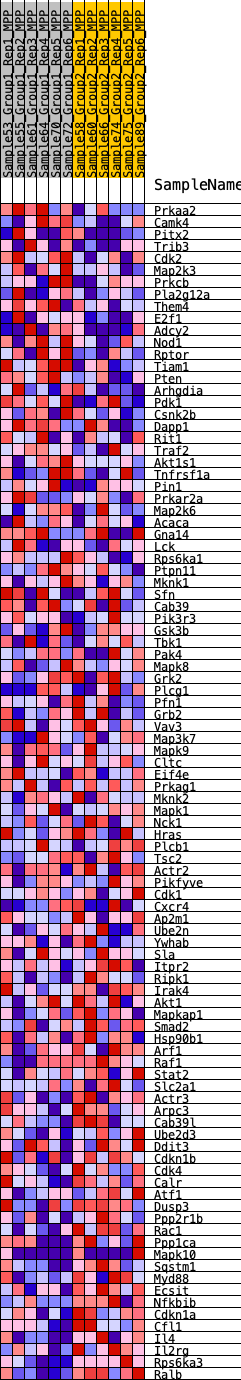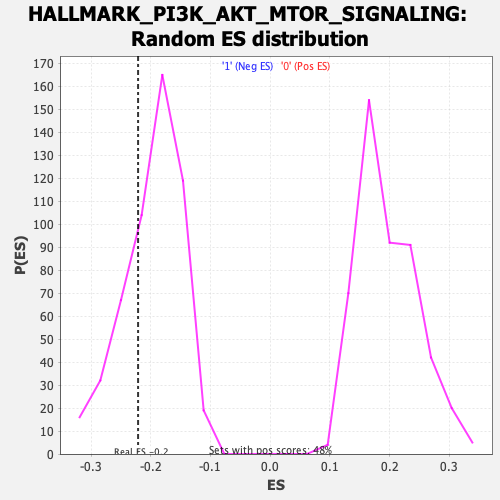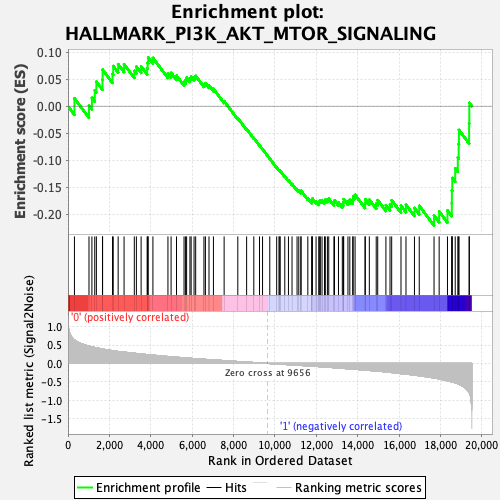
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.22121978 |
| Normalized Enrichment Score (NES) | -1.1240252 |
| Nominal p-value | 0.3045977 |
| FDR q-value | 0.7764044 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkaa2 | 311 | 0.639 | 0.0145 | No |
| 2 | Camk4 | 1014 | 0.475 | 0.0011 | No |
| 3 | Pitx2 | 1155 | 0.454 | 0.0155 | No |
| 4 | Trib3 | 1293 | 0.434 | 0.0292 | No |
| 5 | Cdk2 | 1371 | 0.424 | 0.0455 | No |
| 6 | Map2k3 | 1666 | 0.393 | 0.0492 | No |
| 7 | Prkcb | 1672 | 0.392 | 0.0676 | No |
| 8 | Pla2g12a | 2150 | 0.348 | 0.0597 | No |
| 9 | Them4 | 2185 | 0.345 | 0.0744 | No |
| 10 | E2f1 | 2423 | 0.325 | 0.0778 | No |
| 11 | Adcy2 | 2709 | 0.307 | 0.0777 | No |
| 12 | Nod1 | 3204 | 0.276 | 0.0655 | No |
| 13 | Rptor | 3304 | 0.270 | 0.0733 | No |
| 14 | Tiam1 | 3533 | 0.255 | 0.0737 | No |
| 15 | Pten | 3824 | 0.240 | 0.0703 | No |
| 16 | Arhgdia | 3846 | 0.238 | 0.0806 | No |
| 17 | Pdk1 | 3877 | 0.236 | 0.0903 | No |
| 18 | Csnk2b | 4103 | 0.225 | 0.0894 | No |
| 19 | Dapp1 | 4829 | 0.188 | 0.0611 | No |
| 20 | Rit1 | 4981 | 0.181 | 0.0620 | No |
| 21 | Traf2 | 5243 | 0.169 | 0.0566 | No |
| 22 | Akt1s1 | 5607 | 0.152 | 0.0452 | No |
| 23 | Tnfrsf1a | 5682 | 0.148 | 0.0485 | No |
| 24 | Pin1 | 5731 | 0.146 | 0.0530 | No |
| 25 | Prkar2a | 5892 | 0.140 | 0.0514 | No |
| 26 | Map2k6 | 5950 | 0.137 | 0.0550 | No |
| 27 | Acaca | 6091 | 0.130 | 0.0540 | No |
| 28 | Gna14 | 6169 | 0.127 | 0.0561 | No |
| 29 | Lck | 6569 | 0.113 | 0.0410 | No |
| 30 | Rps6ka1 | 6640 | 0.110 | 0.0426 | No |
| 31 | Ptpn11 | 6813 | 0.103 | 0.0387 | No |
| 32 | Mknk1 | 7027 | 0.096 | 0.0323 | No |
| 33 | Sfn | 7544 | 0.076 | 0.0094 | No |
| 34 | Cab39 | 8205 | 0.051 | -0.0222 | No |
| 35 | Pik3r3 | 8631 | 0.036 | -0.0423 | No |
| 36 | Gsk3b | 8978 | 0.025 | -0.0590 | No |
| 37 | Tbk1 | 9253 | 0.014 | -0.0724 | No |
| 38 | Pak4 | 9400 | 0.009 | -0.0795 | No |
| 39 | Mapk8 | 9750 | -0.003 | -0.0973 | No |
| 40 | Grk2 | 10083 | -0.014 | -0.1137 | No |
| 41 | Plcg1 | 10194 | -0.017 | -0.1186 | No |
| 42 | Pfn1 | 10236 | -0.019 | -0.1198 | No |
| 43 | Grb2 | 10247 | -0.019 | -0.1194 | No |
| 44 | Vav3 | 10475 | -0.027 | -0.1298 | No |
| 45 | Map3k7 | 10652 | -0.033 | -0.1373 | No |
| 46 | Mapk9 | 10825 | -0.039 | -0.1442 | No |
| 47 | Cltc | 11068 | -0.048 | -0.1544 | No |
| 48 | Eif4e | 11151 | -0.050 | -0.1562 | No |
| 49 | Prkag1 | 11252 | -0.054 | -0.1588 | No |
| 50 | Mknk2 | 11260 | -0.054 | -0.1566 | No |
| 51 | Mapk1 | 11588 | -0.065 | -0.1703 | No |
| 52 | Nck1 | 11779 | -0.073 | -0.1766 | No |
| 53 | Hras | 11787 | -0.073 | -0.1735 | No |
| 54 | Plcb1 | 11809 | -0.074 | -0.1710 | No |
| 55 | Tsc2 | 11987 | -0.081 | -0.1763 | No |
| 56 | Actr2 | 12118 | -0.085 | -0.1789 | No |
| 57 | Pikfyve | 12144 | -0.086 | -0.1761 | No |
| 58 | Cdk1 | 12194 | -0.089 | -0.1744 | No |
| 59 | Cxcr4 | 12275 | -0.092 | -0.1741 | No |
| 60 | Ap2m1 | 12393 | -0.097 | -0.1755 | No |
| 61 | Ube2n | 12434 | -0.099 | -0.1728 | No |
| 62 | Ywhab | 12534 | -0.102 | -0.1730 | No |
| 63 | Sla | 12599 | -0.105 | -0.1713 | No |
| 64 | Itpr2 | 12849 | -0.115 | -0.1787 | No |
| 65 | Ripk1 | 12877 | -0.116 | -0.1745 | No |
| 66 | Irak4 | 13069 | -0.124 | -0.1784 | No |
| 67 | Akt1 | 13255 | -0.131 | -0.1817 | No |
| 68 | Mapkap1 | 13306 | -0.133 | -0.1779 | No |
| 69 | Smad2 | 13321 | -0.134 | -0.1722 | No |
| 70 | Hsp90b1 | 13527 | -0.144 | -0.1759 | No |
| 71 | Arf1 | 13617 | -0.148 | -0.1734 | No |
| 72 | Raf1 | 13757 | -0.153 | -0.1733 | No |
| 73 | Stat2 | 13779 | -0.154 | -0.1670 | No |
| 74 | Slc2a1 | 13871 | -0.157 | -0.1642 | No |
| 75 | Actr3 | 14351 | -0.181 | -0.1802 | No |
| 76 | Arpc3 | 14366 | -0.182 | -0.1722 | No |
| 77 | Cab39l | 14560 | -0.190 | -0.1731 | No |
| 78 | Ube2d3 | 14893 | -0.206 | -0.1804 | No |
| 79 | Ddit3 | 14965 | -0.210 | -0.1740 | No |
| 80 | Cdkn1b | 15361 | -0.230 | -0.1833 | No |
| 81 | Cdk4 | 15560 | -0.242 | -0.1820 | No |
| 82 | Calr | 15640 | -0.247 | -0.1742 | No |
| 83 | Atf1 | 16090 | -0.278 | -0.1841 | No |
| 84 | Dusp3 | 16331 | -0.290 | -0.1826 | No |
| 85 | Ppp2r1b | 16740 | -0.317 | -0.1885 | No |
| 86 | Rac1 | 16972 | -0.334 | -0.1844 | No |
| 87 | Ppp1ca | 17687 | -0.394 | -0.2024 | Yes |
| 88 | Mapk10 | 17934 | -0.420 | -0.1950 | Yes |
| 89 | Sqstm1 | 18336 | -0.471 | -0.1932 | Yes |
| 90 | Myd88 | 18545 | -0.501 | -0.1799 | Yes |
| 91 | Ecsit | 18549 | -0.501 | -0.1561 | Yes |
| 92 | Nfkbib | 18568 | -0.504 | -0.1330 | Yes |
| 93 | Cdkn1a | 18710 | -0.528 | -0.1150 | Yes |
| 94 | Cfl1 | 18836 | -0.551 | -0.0952 | Yes |
| 95 | Il4 | 18877 | -0.562 | -0.0704 | Yes |
| 96 | Il2rg | 18890 | -0.565 | -0.0440 | Yes |
| 97 | Rps6ka3 | 19382 | -0.785 | -0.0318 | Yes |
| 98 | Ralb | 19392 | -0.804 | 0.0062 | Yes |