Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
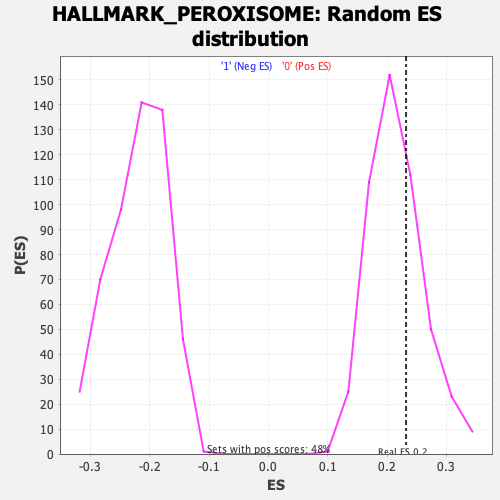
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.23194703 |
| Normalized Enrichment Score (NES) | 1.0731646 |
| Nominal p-value | 0.32016632 |
| FDR q-value | 0.7824586 |
| FWER p-Value | 0.981 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd3b7 | 65 | 0.864 | 0.0498 | Yes |
| 2 | Abcd2 | 214 | 0.691 | 0.0847 | Yes |
| 3 | Ehhadh | 668 | 0.533 | 0.0941 | Yes |
| 4 | Pex2 | 926 | 0.487 | 0.1109 | Yes |
| 5 | Gstk1 | 1047 | 0.470 | 0.1336 | Yes |
| 6 | Lonp2 | 2203 | 0.343 | 0.0952 | Yes |
| 7 | Mlycd | 2683 | 0.308 | 0.0895 | Yes |
| 8 | Fdps | 2898 | 0.296 | 0.0967 | Yes |
| 9 | Slc25a17 | 3294 | 0.271 | 0.0930 | Yes |
| 10 | Ercc3 | 3365 | 0.266 | 0.1058 | Yes |
| 11 | Acsl1 | 3398 | 0.264 | 0.1204 | Yes |
| 12 | Idi1 | 3681 | 0.248 | 0.1211 | Yes |
| 13 | Abcb1a | 3698 | 0.247 | 0.1355 | Yes |
| 14 | Mvp | 3818 | 0.240 | 0.1441 | Yes |
| 15 | Abcb4 | 3979 | 0.233 | 0.1502 | Yes |
| 16 | Cacna1b | 4033 | 0.229 | 0.1615 | Yes |
| 17 | Prdx5 | 4162 | 0.222 | 0.1686 | Yes |
| 18 | Fads1 | 4169 | 0.221 | 0.1819 | Yes |
| 19 | Gnpat | 4174 | 0.221 | 0.1953 | Yes |
| 20 | Siah1a | 4180 | 0.221 | 0.2086 | Yes |
| 21 | Sult2b1 | 4227 | 0.219 | 0.2197 | Yes |
| 22 | Abcd1 | 4429 | 0.208 | 0.2222 | Yes |
| 23 | Abcc5 | 4702 | 0.193 | 0.2201 | Yes |
| 24 | Slc25a19 | 4855 | 0.187 | 0.2238 | Yes |
| 25 | Eci2 | 5071 | 0.177 | 0.2236 | Yes |
| 26 | Sema3c | 5382 | 0.163 | 0.2176 | Yes |
| 27 | Slc23a2 | 5502 | 0.157 | 0.2211 | Yes |
| 28 | Aldh9a1 | 5670 | 0.149 | 0.2217 | Yes |
| 29 | Ide | 5689 | 0.148 | 0.2299 | Yes |
| 30 | Ephx2 | 5820 | 0.142 | 0.2319 | Yes |
| 31 | Smarcc1 | 6845 | 0.102 | 0.1855 | No |
| 32 | Hsd17b4 | 7287 | 0.086 | 0.1681 | No |
| 33 | Pex14 | 7399 | 0.082 | 0.1674 | No |
| 34 | Pex6 | 7603 | 0.074 | 0.1615 | No |
| 35 | Msh2 | 7716 | 0.070 | 0.1600 | No |
| 36 | Cnbp | 8348 | 0.046 | 0.1304 | No |
| 37 | Cat | 8398 | 0.044 | 0.1306 | No |
| 38 | Crat | 8465 | 0.042 | 0.1297 | No |
| 39 | Slc27a2 | 8538 | 0.040 | 0.1285 | No |
| 40 | Abcb9 | 8940 | 0.026 | 0.1094 | No |
| 41 | Dhcr24 | 8967 | 0.025 | 0.1096 | No |
| 42 | Acaa1a | 9068 | 0.022 | 0.1058 | No |
| 43 | Isoc1 | 9086 | 0.021 | 0.1062 | No |
| 44 | Hsd17b11 | 9236 | 0.015 | 0.0995 | No |
| 45 | Acsl4 | 9362 | 0.011 | 0.0937 | No |
| 46 | Hmgcl | 9552 | 0.004 | 0.0842 | No |
| 47 | Cdk7 | 9773 | -0.004 | 0.0731 | No |
| 48 | Bcl10 | 9831 | -0.006 | 0.0705 | No |
| 49 | Ywhah | 9859 | -0.007 | 0.0695 | No |
| 50 | Ctbp1 | 10046 | -0.013 | 0.0607 | No |
| 51 | Aldh1a1 | 10066 | -0.014 | 0.0606 | No |
| 52 | Pabpc1 | 10134 | -0.016 | 0.0581 | No |
| 53 | Abcd3 | 10261 | -0.019 | 0.0528 | No |
| 54 | Dlg4 | 10315 | -0.022 | 0.0514 | No |
| 55 | Fis1 | 10408 | -0.025 | 0.0483 | No |
| 56 | Dhrs3 | 10413 | -0.025 | 0.0496 | No |
| 57 | Acox1 | 10459 | -0.027 | 0.0489 | No |
| 58 | Pex5 | 10509 | -0.028 | 0.0482 | No |
| 59 | Pex11a | 10872 | -0.041 | 0.0320 | No |
| 60 | Acsl5 | 11019 | -0.046 | 0.0273 | No |
| 61 | Esr2 | 11025 | -0.046 | 0.0299 | No |
| 62 | Cadm1 | 11299 | -0.056 | 0.0193 | No |
| 63 | Hras | 11787 | -0.073 | -0.0013 | No |
| 64 | Vps4b | 12072 | -0.083 | -0.0107 | No |
| 65 | Ech1 | 12090 | -0.084 | -0.0064 | No |
| 66 | Acot8 | 12507 | -0.101 | -0.0216 | No |
| 67 | Hsd11b2 | 13190 | -0.129 | -0.0488 | No |
| 68 | Sod2 | 13375 | -0.138 | -0.0498 | No |
| 69 | Idh2 | 13419 | -0.139 | -0.0434 | No |
| 70 | Ercc1 | 13861 | -0.157 | -0.0565 | No |
| 71 | Atxn1 | 14046 | -0.167 | -0.0557 | No |
| 72 | Elovl5 | 14416 | -0.184 | -0.0634 | No |
| 73 | Itgb1bp1 | 14530 | -0.189 | -0.0576 | No |
| 74 | Rdh11 | 14554 | -0.190 | -0.0471 | No |
| 75 | Slc35b2 | 14835 | -0.203 | -0.0490 | No |
| 76 | Cln8 | 14926 | -0.208 | -0.0408 | No |
| 77 | Idh1 | 15246 | -0.223 | -0.0435 | No |
| 78 | Sod1 | 15858 | -0.261 | -0.0589 | No |
| 79 | Cln6 | 15869 | -0.262 | -0.0433 | No |
| 80 | Prdx1 | 16163 | -0.282 | -0.0410 | No |
| 81 | Top2a | 16758 | -0.318 | -0.0521 | No |
| 82 | Scp2 | 17055 | -0.340 | -0.0464 | No |
| 83 | Retsat | 17058 | -0.340 | -0.0256 | No |
| 84 | Nudt19 | 17498 | -0.378 | -0.0249 | No |
| 85 | Pex13 | 18288 | -0.465 | -0.0369 | No |
| 86 | Pex11b | 18304 | -0.466 | -0.0090 | No |
| 87 | Tspo | 18909 | -0.570 | -0.0051 | No |
| 88 | Slc25a4 | 18962 | -0.586 | 0.0283 | No |