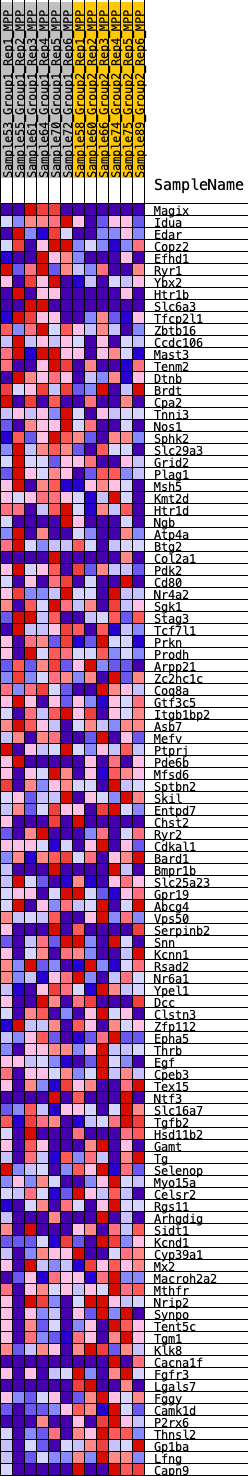
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
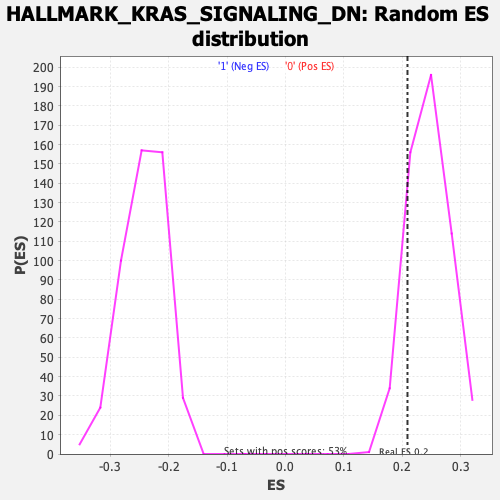
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.20913751 |
| Normalized Enrichment Score (NES) | 0.8517518 |
| Nominal p-value | 0.8336484 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Magix | 409 | 0.598 | 0.0019 | Yes |
| 2 | Idua | 565 | 0.552 | 0.0151 | Yes |
| 3 | Edar | 947 | 0.484 | 0.0140 | Yes |
| 4 | Copz2 | 996 | 0.477 | 0.0298 | Yes |
| 5 | Efhd1 | 1078 | 0.464 | 0.0435 | Yes |
| 6 | Ryr1 | 1219 | 0.443 | 0.0532 | Yes |
| 7 | Ybx2 | 1235 | 0.441 | 0.0694 | Yes |
| 8 | Htr1b | 1323 | 0.430 | 0.0814 | Yes |
| 9 | Slc6a3 | 1420 | 0.419 | 0.0926 | Yes |
| 10 | Tfcp2l1 | 1546 | 0.406 | 0.1017 | Yes |
| 11 | Zbtb16 | 1591 | 0.400 | 0.1148 | Yes |
| 12 | Ccdc106 | 1756 | 0.384 | 0.1211 | Yes |
| 13 | Mast3 | 1828 | 0.376 | 0.1318 | Yes |
| 14 | Tenm2 | 1965 | 0.363 | 0.1387 | Yes |
| 15 | Dtnb | 2111 | 0.351 | 0.1447 | Yes |
| 16 | Brdt | 2209 | 0.342 | 0.1529 | Yes |
| 17 | Cpa2 | 2237 | 0.339 | 0.1645 | Yes |
| 18 | Tnni3 | 2238 | 0.339 | 0.1775 | Yes |
| 19 | Nos1 | 2266 | 0.337 | 0.1891 | Yes |
| 20 | Sphk2 | 2325 | 0.332 | 0.1988 | Yes |
| 21 | Slc29a3 | 2597 | 0.314 | 0.1969 | Yes |
| 22 | Grid2 | 3037 | 0.286 | 0.1853 | Yes |
| 23 | Plag1 | 3240 | 0.275 | 0.1854 | Yes |
| 24 | Msh5 | 3320 | 0.269 | 0.1917 | Yes |
| 25 | Kmt2d | 3432 | 0.262 | 0.1960 | Yes |
| 26 | Htr1d | 3434 | 0.261 | 0.2060 | Yes |
| 27 | Ngb | 3670 | 0.248 | 0.2034 | Yes |
| 28 | Atp4a | 3741 | 0.244 | 0.2091 | Yes |
| 29 | Btg2 | 4231 | 0.219 | 0.1923 | No |
| 30 | Col2a1 | 4350 | 0.213 | 0.1944 | No |
| 31 | Pdk2 | 4732 | 0.192 | 0.1821 | No |
| 32 | Cd80 | 4774 | 0.191 | 0.1873 | No |
| 33 | Nr4a2 | 4818 | 0.189 | 0.1924 | No |
| 34 | Sgk1 | 4872 | 0.187 | 0.1968 | No |
| 35 | Stag3 | 5205 | 0.171 | 0.1863 | No |
| 36 | Tcf7l1 | 5418 | 0.161 | 0.1815 | No |
| 37 | Prkn | 5467 | 0.158 | 0.1851 | No |
| 38 | Prodh | 5800 | 0.143 | 0.1735 | No |
| 39 | Arpp21 | 5933 | 0.138 | 0.1720 | No |
| 40 | Zc2hc1c | 5995 | 0.135 | 0.1740 | No |
| 41 | Coq8a | 6255 | 0.123 | 0.1654 | No |
| 42 | Gtf3c5 | 6687 | 0.108 | 0.1473 | No |
| 43 | Itgb1bp2 | 7374 | 0.083 | 0.1152 | No |
| 44 | Asb7 | 7835 | 0.066 | 0.0940 | No |
| 45 | Mefv | 8041 | 0.058 | 0.0857 | No |
| 46 | Ptprj | 8258 | 0.049 | 0.0764 | No |
| 47 | Pde6b | 8358 | 0.045 | 0.0731 | No |
| 48 | Mfsd6 | 8378 | 0.045 | 0.0738 | No |
| 49 | Sptbn2 | 8463 | 0.042 | 0.0711 | No |
| 50 | Skil | 9234 | 0.015 | 0.0320 | No |
| 51 | Entpd7 | 9422 | 0.008 | 0.0227 | No |
| 52 | Chst2 | 9462 | 0.007 | 0.0209 | No |
| 53 | Ryr2 | 9696 | -0.001 | 0.0090 | No |
| 54 | Cdkal1 | 9866 | -0.007 | 0.0005 | No |
| 55 | Bard1 | 9991 | -0.011 | -0.0055 | No |
| 56 | Bmpr1b | 10024 | -0.012 | -0.0066 | No |
| 57 | Slc25a23 | 10088 | -0.015 | -0.0093 | No |
| 58 | Gpr19 | 10450 | -0.026 | -0.0269 | No |
| 59 | Abcg4 | 10666 | -0.034 | -0.0367 | No |
| 60 | Vps50 | 10683 | -0.035 | -0.0362 | No |
| 61 | Serpinb2 | 10935 | -0.043 | -0.0475 | No |
| 62 | Snn | 11060 | -0.047 | -0.0521 | No |
| 63 | Kcnn1 | 11064 | -0.048 | -0.0504 | No |
| 64 | Rsad2 | 11301 | -0.056 | -0.0604 | No |
| 65 | Nr6a1 | 11422 | -0.060 | -0.0643 | No |
| 66 | Ypel1 | 11673 | -0.069 | -0.0745 | No |
| 67 | Dcc | 11838 | -0.075 | -0.0801 | No |
| 68 | Clstn3 | 11951 | -0.079 | -0.0829 | No |
| 69 | Zfp112 | 12398 | -0.097 | -0.1021 | No |
| 70 | Epha5 | 12495 | -0.101 | -0.1032 | No |
| 71 | Thrb | 12538 | -0.102 | -0.1014 | No |
| 72 | Egf | 12595 | -0.104 | -0.1003 | No |
| 73 | Cpeb3 | 12752 | -0.111 | -0.1041 | No |
| 74 | Tex15 | 12789 | -0.113 | -0.1016 | No |
| 75 | Ntf3 | 12865 | -0.115 | -0.1011 | No |
| 76 | Slc16a7 | 12941 | -0.119 | -0.1004 | No |
| 77 | Tgfb2 | 13048 | -0.123 | -0.1011 | No |
| 78 | Hsd11b2 | 13190 | -0.129 | -0.1034 | No |
| 79 | Gamt | 13307 | -0.133 | -0.1043 | No |
| 80 | Tg | 14019 | -0.165 | -0.1346 | No |
| 81 | Selenop | 14327 | -0.180 | -0.1435 | No |
| 82 | Myo15a | 14364 | -0.182 | -0.1384 | No |
| 83 | Celsr2 | 15134 | -0.218 | -0.1696 | No |
| 84 | Rgs11 | 15274 | -0.225 | -0.1682 | No |
| 85 | Arhgdig | 15367 | -0.230 | -0.1641 | No |
| 86 | Sidt1 | 15419 | -0.233 | -0.1578 | No |
| 87 | Kcnd1 | 15609 | -0.245 | -0.1581 | No |
| 88 | Cyp39a1 | 15776 | -0.256 | -0.1568 | No |
| 89 | Mx2 | 16329 | -0.290 | -0.1741 | No |
| 90 | Macroh2a2 | 16398 | -0.296 | -0.1663 | No |
| 91 | Mthfr | 16633 | -0.311 | -0.1664 | No |
| 92 | Nrip2 | 16794 | -0.321 | -0.1624 | No |
| 93 | Synpo | 17437 | -0.372 | -0.1812 | No |
| 94 | Tent5c | 17492 | -0.377 | -0.1695 | No |
| 95 | Tgm1 | 17662 | -0.393 | -0.1631 | No |
| 96 | Klk8 | 17888 | -0.415 | -0.1588 | No |
| 97 | Cacna1f | 17974 | -0.426 | -0.1468 | No |
| 98 | Fgfr3 | 18091 | -0.440 | -0.1360 | No |
| 99 | Lgals7 | 18116 | -0.444 | -0.1202 | No |
| 100 | Fggy | 18401 | -0.479 | -0.1164 | No |
| 101 | Camk1d | 18932 | -0.577 | -0.1216 | No |
| 102 | P2rx6 | 19118 | -0.638 | -0.1066 | No |
| 103 | Thnsl2 | 19346 | -0.753 | -0.0894 | No |
| 104 | Gp1ba | 19375 | -0.777 | -0.0611 | No |
| 105 | Lfng | 19417 | -0.836 | -0.0311 | No |
| 106 | Capn9 | 19457 | -0.937 | 0.0028 | No |