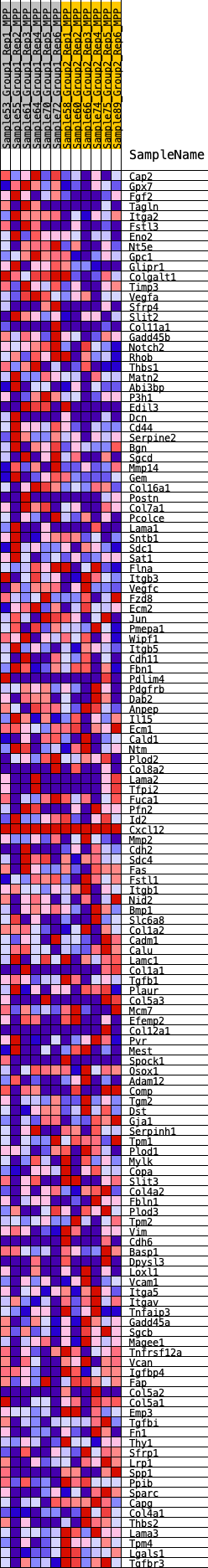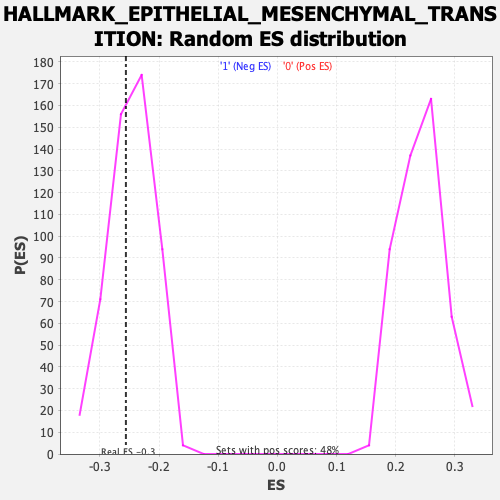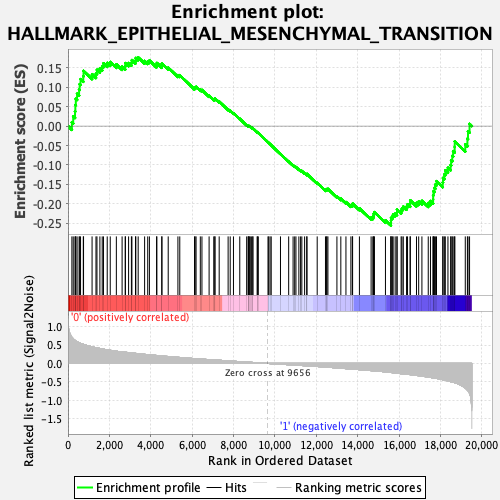
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.25612432 |
| Normalized Enrichment Score (NES) | -1.0432287 |
| Nominal p-value | 0.35589942 |
| FDR q-value | 0.8774369 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cap2 | 188 | 0.709 | 0.0099 | No |
| 2 | Gpx7 | 252 | 0.670 | 0.0252 | No |
| 3 | Fgf2 | 340 | 0.625 | 0.0381 | No |
| 4 | Tagln | 355 | 0.618 | 0.0545 | No |
| 5 | Itga2 | 378 | 0.609 | 0.0702 | No |
| 6 | Fstl3 | 439 | 0.588 | 0.0834 | No |
| 7 | Eno2 | 536 | 0.559 | 0.0939 | No |
| 8 | Nt5e | 563 | 0.552 | 0.1079 | No |
| 9 | Gpc1 | 606 | 0.544 | 0.1208 | No |
| 10 | Glipr1 | 739 | 0.519 | 0.1283 | No |
| 11 | Colgalt1 | 751 | 0.517 | 0.1421 | No |
| 12 | Timp3 | 1163 | 0.453 | 0.1334 | No |
| 13 | Vegfa | 1352 | 0.426 | 0.1355 | No |
| 14 | Sfrp4 | 1399 | 0.421 | 0.1448 | No |
| 15 | Slit2 | 1541 | 0.407 | 0.1488 | No |
| 16 | Col11a1 | 1653 | 0.394 | 0.1540 | No |
| 17 | Gadd45b | 1714 | 0.388 | 0.1616 | No |
| 18 | Notch2 | 1895 | 0.369 | 0.1626 | No |
| 19 | Rhob | 2041 | 0.357 | 0.1650 | No |
| 20 | Thbs1 | 2336 | 0.331 | 0.1590 | No |
| 21 | Matn2 | 2614 | 0.313 | 0.1533 | No |
| 22 | Abi3bp | 2766 | 0.305 | 0.1540 | No |
| 23 | P3h1 | 2773 | 0.304 | 0.1621 | No |
| 24 | Edil3 | 2928 | 0.294 | 0.1623 | No |
| 25 | Dcn | 3063 | 0.285 | 0.1632 | No |
| 26 | Cd44 | 3088 | 0.283 | 0.1698 | No |
| 27 | Serpine2 | 3261 | 0.273 | 0.1685 | No |
| 28 | Bgn | 3277 | 0.272 | 0.1753 | No |
| 29 | Sgcd | 3391 | 0.265 | 0.1768 | No |
| 30 | Mmp14 | 3702 | 0.247 | 0.1676 | No |
| 31 | Gem | 3852 | 0.238 | 0.1665 | No |
| 32 | Col16a1 | 3933 | 0.235 | 0.1689 | No |
| 33 | Postn | 4280 | 0.216 | 0.1570 | No |
| 34 | Col7a1 | 4293 | 0.216 | 0.1624 | No |
| 35 | Pcolce | 4533 | 0.203 | 0.1557 | No |
| 36 | Lama1 | 4537 | 0.203 | 0.1611 | No |
| 37 | Sntb1 | 4842 | 0.188 | 0.1506 | No |
| 38 | Sdc1 | 5311 | 0.166 | 0.1311 | No |
| 39 | Sat1 | 5401 | 0.162 | 0.1310 | No |
| 40 | Flna | 6113 | 0.129 | 0.0978 | No |
| 41 | Itgb3 | 6141 | 0.128 | 0.1000 | No |
| 42 | Vegfc | 6188 | 0.126 | 0.1011 | No |
| 43 | Fzd8 | 6393 | 0.118 | 0.0938 | No |
| 44 | Ecm2 | 6472 | 0.115 | 0.0930 | No |
| 45 | Jun | 6818 | 0.103 | 0.0781 | No |
| 46 | Pmepa1 | 7043 | 0.095 | 0.0691 | No |
| 47 | Wipf1 | 7095 | 0.093 | 0.0691 | No |
| 48 | Itgb5 | 7101 | 0.093 | 0.0714 | No |
| 49 | Cdh11 | 7305 | 0.085 | 0.0633 | No |
| 50 | Fbn1 | 7737 | 0.069 | 0.0430 | No |
| 51 | Pdlim4 | 7838 | 0.065 | 0.0396 | No |
| 52 | Pdgfrb | 7995 | 0.060 | 0.0332 | No |
| 53 | Dab2 | 8300 | 0.047 | 0.0189 | No |
| 54 | Anpep | 8633 | 0.036 | 0.0027 | No |
| 55 | Il15 | 8704 | 0.034 | 0.0001 | No |
| 56 | Ecm1 | 8718 | 0.034 | 0.0003 | No |
| 57 | Cald1 | 8741 | 0.033 | 0.0001 | No |
| 58 | Ntm | 8762 | 0.032 | -0.0000 | No |
| 59 | Plod2 | 8807 | 0.031 | -0.0014 | No |
| 60 | Col8a2 | 8861 | 0.029 | -0.0034 | No |
| 61 | Lama2 | 8931 | 0.026 | -0.0062 | No |
| 62 | Tfpi2 | 8932 | 0.026 | -0.0055 | No |
| 63 | Fuca1 | 9145 | 0.018 | -0.0159 | No |
| 64 | Pfn2 | 9148 | 0.018 | -0.0155 | No |
| 65 | Id2 | 9196 | 0.017 | -0.0175 | No |
| 66 | Cxcl12 | 9664 | 0.000 | -0.0416 | No |
| 67 | Mmp2 | 9697 | -0.001 | -0.0432 | No |
| 68 | Cdh2 | 9763 | -0.003 | -0.0465 | No |
| 69 | Sdc4 | 9819 | -0.005 | -0.0492 | No |
| 70 | Fas | 10267 | -0.020 | -0.0717 | No |
| 71 | Fstl1 | 10663 | -0.034 | -0.0912 | No |
| 72 | Itgb1 | 10876 | -0.041 | -0.1010 | No |
| 73 | Nid2 | 10953 | -0.044 | -0.1037 | No |
| 74 | Bmp1 | 11014 | -0.046 | -0.1055 | No |
| 75 | Slc6a8 | 11155 | -0.051 | -0.1113 | No |
| 76 | Col1a2 | 11239 | -0.053 | -0.1142 | No |
| 77 | Cadm1 | 11299 | -0.056 | -0.1157 | No |
| 78 | Calu | 11433 | -0.060 | -0.1209 | No |
| 79 | Lamc1 | 11537 | -0.064 | -0.1244 | No |
| 80 | Col1a1 | 11546 | -0.064 | -0.1231 | No |
| 81 | Tgfb1 | 12041 | -0.082 | -0.1463 | No |
| 82 | Plaur | 12441 | -0.099 | -0.1641 | No |
| 83 | Col5a3 | 12484 | -0.100 | -0.1635 | No |
| 84 | Mcm7 | 12511 | -0.101 | -0.1621 | No |
| 85 | Efemp2 | 12559 | -0.103 | -0.1616 | No |
| 86 | Col12a1 | 12994 | -0.122 | -0.1807 | No |
| 87 | Pvr | 13183 | -0.129 | -0.1868 | No |
| 88 | Mest | 13430 | -0.140 | -0.1956 | No |
| 89 | Spock1 | 13669 | -0.150 | -0.2037 | No |
| 90 | Qsox1 | 13740 | -0.152 | -0.2032 | No |
| 91 | Adam12 | 13752 | -0.152 | -0.1995 | No |
| 92 | Comp | 14081 | -0.168 | -0.2118 | No |
| 93 | Tgm2 | 14646 | -0.194 | -0.2355 | No |
| 94 | Dst | 14729 | -0.198 | -0.2343 | No |
| 95 | Gja1 | 14764 | -0.199 | -0.2305 | No |
| 96 | Serpinh1 | 14775 | -0.200 | -0.2255 | No |
| 97 | Tpm1 | 14807 | -0.201 | -0.2215 | No |
| 98 | Plod1 | 15339 | -0.229 | -0.2425 | No |
| 99 | Mylk | 15603 | -0.245 | -0.2493 | Yes |
| 100 | Copa | 15606 | -0.245 | -0.2427 | Yes |
| 101 | Slit3 | 15611 | -0.246 | -0.2361 | Yes |
| 102 | Col4a2 | 15672 | -0.250 | -0.2322 | Yes |
| 103 | Fbln1 | 15710 | -0.252 | -0.2272 | Yes |
| 104 | Plod3 | 15808 | -0.258 | -0.2250 | Yes |
| 105 | Tpm2 | 15897 | -0.264 | -0.2222 | Yes |
| 106 | Vim | 15898 | -0.264 | -0.2149 | Yes |
| 107 | Cdh6 | 16096 | -0.278 | -0.2174 | Yes |
| 108 | Basp1 | 16137 | -0.280 | -0.2117 | Yes |
| 109 | Dpysl3 | 16201 | -0.284 | -0.2071 | Yes |
| 110 | Loxl1 | 16359 | -0.292 | -0.2071 | Yes |
| 111 | Vcam1 | 16406 | -0.296 | -0.2013 | Yes |
| 112 | Itga5 | 16533 | -0.303 | -0.1994 | Yes |
| 113 | Itgav | 16538 | -0.303 | -0.1912 | Yes |
| 114 | Tnfaip3 | 16839 | -0.324 | -0.1977 | Yes |
| 115 | Gadd45a | 16945 | -0.332 | -0.1940 | Yes |
| 116 | Sgcb | 17101 | -0.344 | -0.1924 | Yes |
| 117 | Magee1 | 17407 | -0.370 | -0.1979 | Yes |
| 118 | Tnfrsf12a | 17514 | -0.380 | -0.1929 | Yes |
| 119 | Vcan | 17642 | -0.391 | -0.1886 | Yes |
| 120 | Igfbp4 | 17645 | -0.392 | -0.1779 | Yes |
| 121 | Fap | 17660 | -0.392 | -0.1677 | Yes |
| 122 | Col5a2 | 17706 | -0.396 | -0.1591 | Yes |
| 123 | Col5a1 | 17752 | -0.401 | -0.1503 | Yes |
| 124 | Emp3 | 17806 | -0.406 | -0.1418 | Yes |
| 125 | Tgfbi | 18108 | -0.442 | -0.1451 | Yes |
| 126 | Fn1 | 18121 | -0.444 | -0.1334 | Yes |
| 127 | Thy1 | 18185 | -0.450 | -0.1242 | Yes |
| 128 | Sfrp1 | 18231 | -0.457 | -0.1139 | Yes |
| 129 | Lrp1 | 18359 | -0.473 | -0.1073 | Yes |
| 130 | Spp1 | 18489 | -0.494 | -0.1003 | Yes |
| 131 | Ppib | 18514 | -0.498 | -0.0877 | Yes |
| 132 | Sparc | 18569 | -0.504 | -0.0766 | Yes |
| 133 | Capg | 18610 | -0.511 | -0.0645 | Yes |
| 134 | Col4a1 | 18684 | -0.523 | -0.0538 | Yes |
| 135 | Thbs2 | 18688 | -0.523 | -0.0394 | Yes |
| 136 | Lama3 | 19196 | -0.668 | -0.0471 | Yes |
| 137 | Tpm4 | 19299 | -0.725 | -0.0323 | Yes |
| 138 | Lgals1 | 19328 | -0.739 | -0.0132 | Yes |
| 139 | Tgfbr3 | 19403 | -0.819 | 0.0056 | Yes |