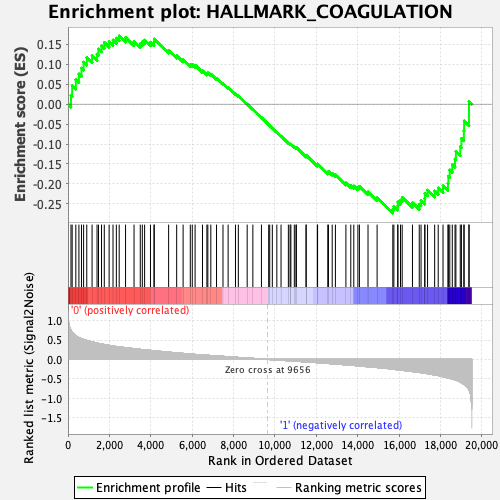
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.27326256 |
| Normalized Enrichment Score (NES) | -1.1164511 |
| Nominal p-value | 0.26260504 |
| FDR q-value | 0.74410087 |
| FWER p-Value | 0.978 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plek | 144 | 0.750 | 0.0220 | No |
| 2 | Msrb2 | 201 | 0.700 | 0.0466 | No |
| 3 | Itga2 | 378 | 0.609 | 0.0614 | No |
| 4 | Pdgfb | 527 | 0.561 | 0.0758 | No |
| 5 | Sh2b2 | 651 | 0.537 | 0.0906 | No |
| 6 | Gsn | 754 | 0.517 | 0.1056 | No |
| 7 | Masp2 | 907 | 0.489 | 0.1170 | No |
| 8 | Timp3 | 1163 | 0.453 | 0.1216 | No |
| 9 | Ang | 1403 | 0.421 | 0.1258 | No |
| 10 | Dusp6 | 1474 | 0.413 | 0.1384 | No |
| 11 | Hnf4a | 1620 | 0.398 | 0.1465 | No |
| 12 | Gng12 | 1753 | 0.384 | 0.1548 | No |
| 13 | Rabif | 1987 | 0.362 | 0.1570 | No |
| 14 | Cpn1 | 2175 | 0.346 | 0.1609 | No |
| 15 | Thbs1 | 2336 | 0.331 | 0.1657 | No |
| 16 | Crip2 | 2473 | 0.321 | 0.1712 | No |
| 17 | Mmp11 | 2781 | 0.303 | 0.1673 | No |
| 18 | C2 | 3191 | 0.277 | 0.1571 | No |
| 19 | Capn5 | 3492 | 0.258 | 0.1518 | No |
| 20 | Arf4 | 3591 | 0.252 | 0.1567 | No |
| 21 | Mmp14 | 3702 | 0.247 | 0.1607 | No |
| 22 | Mst1 | 3985 | 0.232 | 0.1553 | No |
| 23 | F8 | 4156 | 0.222 | 0.1552 | No |
| 24 | P2ry1 | 4167 | 0.221 | 0.1634 | No |
| 25 | Ctso | 4862 | 0.187 | 0.1350 | No |
| 26 | Klkb1 | 5251 | 0.169 | 0.1216 | No |
| 27 | Anxa1 | 5562 | 0.154 | 0.1117 | No |
| 28 | Wdr1 | 5909 | 0.139 | 0.0993 | No |
| 29 | C8g | 6005 | 0.134 | 0.0997 | No |
| 30 | Itgb3 | 6141 | 0.128 | 0.0978 | No |
| 31 | Prss23 | 6498 | 0.115 | 0.0839 | No |
| 32 | Gp9 | 6711 | 0.107 | 0.0772 | No |
| 33 | Ctsl | 6757 | 0.105 | 0.0790 | No |
| 34 | Furin | 6901 | 0.100 | 0.0756 | No |
| 35 | Cfi | 7177 | 0.090 | 0.0650 | No |
| 36 | Olr1 | 7489 | 0.079 | 0.0520 | No |
| 37 | Fbn1 | 7737 | 0.069 | 0.0420 | No |
| 38 | Hpn | 8092 | 0.056 | 0.0260 | No |
| 39 | Ctsk | 8230 | 0.050 | 0.0209 | No |
| 40 | Pef1 | 8658 | 0.036 | 0.0003 | No |
| 41 | Tfpi2 | 8932 | 0.026 | -0.0127 | No |
| 42 | Prep | 9347 | 0.011 | -0.0336 | No |
| 43 | Mmp2 | 9697 | -0.001 | -0.0515 | No |
| 44 | S100a1 | 9747 | -0.003 | -0.0539 | No |
| 45 | Usp11 | 9873 | -0.007 | -0.0601 | No |
| 46 | Ctse | 10091 | -0.015 | -0.0707 | No |
| 47 | Lta4h | 10294 | -0.020 | -0.0803 | No |
| 48 | Trf | 10648 | -0.033 | -0.0972 | No |
| 49 | Casp9 | 10730 | -0.036 | -0.0999 | No |
| 50 | Gnb2 | 10769 | -0.037 | -0.1004 | No |
| 51 | Serpinb2 | 10935 | -0.043 | -0.1072 | No |
| 52 | Bmp1 | 11014 | -0.046 | -0.1094 | No |
| 53 | Vwf | 11036 | -0.047 | -0.1087 | No |
| 54 | Dpp4 | 11503 | -0.062 | -0.1302 | No |
| 55 | Serpinc1 | 11507 | -0.063 | -0.1279 | No |
| 56 | F10 | 12045 | -0.082 | -0.1524 | No |
| 57 | Maff | 12060 | -0.083 | -0.1498 | No |
| 58 | Cd9 | 12553 | -0.103 | -0.1711 | No |
| 59 | C9 | 12585 | -0.104 | -0.1686 | No |
| 60 | Fyn | 12766 | -0.111 | -0.1735 | No |
| 61 | Ctsh | 12921 | -0.118 | -0.1768 | No |
| 62 | Iscu | 13428 | -0.140 | -0.1974 | No |
| 63 | C8b | 13664 | -0.150 | -0.2036 | No |
| 64 | Thbd | 13810 | -0.155 | -0.2050 | No |
| 65 | Cpq | 14007 | -0.165 | -0.2087 | No |
| 66 | Comp | 14081 | -0.168 | -0.2058 | No |
| 67 | Clu | 14497 | -0.187 | -0.2198 | No |
| 68 | Csrp1 | 14936 | -0.208 | -0.2342 | No |
| 69 | Lamp2 | 15695 | -0.251 | -0.2634 | Yes |
| 70 | Cfb | 15746 | -0.254 | -0.2560 | Yes |
| 71 | C3 | 15933 | -0.267 | -0.2551 | Yes |
| 72 | S100a13 | 15941 | -0.268 | -0.2450 | Yes |
| 73 | Pf4 | 16067 | -0.276 | -0.2406 | Yes |
| 74 | Adam9 | 16148 | -0.281 | -0.2337 | Yes |
| 75 | Sirt2 | 16644 | -0.312 | -0.2470 | Yes |
| 76 | Rac1 | 16972 | -0.334 | -0.2507 | Yes |
| 77 | F2rl2 | 17059 | -0.340 | -0.2418 | Yes |
| 78 | Plau | 17238 | -0.356 | -0.2370 | Yes |
| 79 | Pecam1 | 17245 | -0.357 | -0.2233 | Yes |
| 80 | Proz | 17374 | -0.368 | -0.2155 | Yes |
| 81 | Cfd | 17717 | -0.397 | -0.2175 | Yes |
| 82 | Klk8 | 17888 | -0.415 | -0.2100 | Yes |
| 83 | Fn1 | 18121 | -0.444 | -0.2045 | Yes |
| 84 | Lrp1 | 18359 | -0.473 | -0.1982 | Yes |
| 85 | Ctsb | 18379 | -0.476 | -0.1805 | Yes |
| 86 | C1qa | 18448 | -0.486 | -0.1649 | Yes |
| 87 | Sparc | 18569 | -0.504 | -0.1513 | Yes |
| 88 | Mmp15 | 18701 | -0.526 | -0.1374 | Yes |
| 89 | Pros1 | 18743 | -0.533 | -0.1186 | Yes |
| 90 | Klf7 | 18954 | -0.583 | -0.1066 | Yes |
| 91 | Rapgef3 | 19010 | -0.599 | -0.0859 | Yes |
| 92 | Lgmn | 19129 | -0.641 | -0.0668 | Yes |
| 93 | Gda | 19140 | -0.645 | -0.0420 | Yes |
| 94 | Gp1ba | 19375 | -0.777 | -0.0236 | Yes |
| 95 | Capn2 | 19377 | -0.780 | 0.0070 | Yes |

