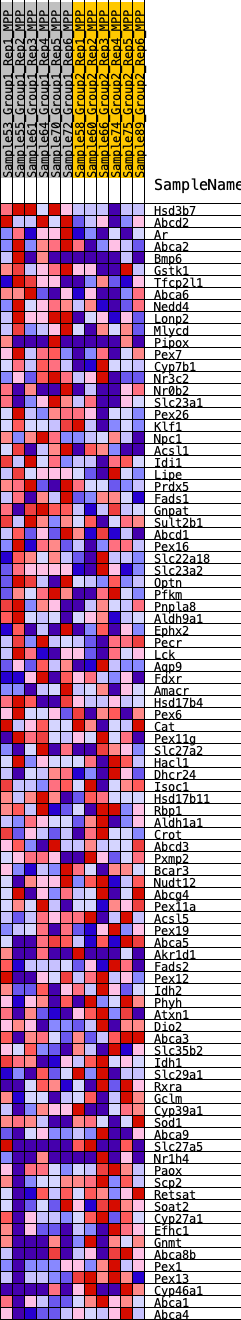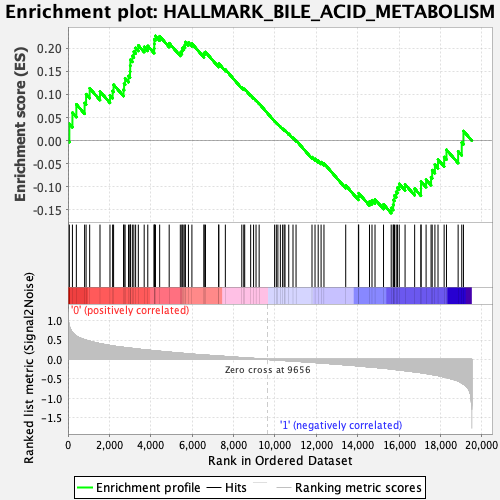
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MPP.MPP_Pheno.cls#Group1_versus_Group2.MPP_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | MPP_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.2266346 |
| Normalized Enrichment Score (NES) | 0.9311055 |
| Nominal p-value | 0.56496066 |
| FDR q-value | 0.93549496 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd3b7 | 65 | 0.864 | 0.0364 | Yes |
| 2 | Abcd2 | 214 | 0.691 | 0.0606 | Yes |
| 3 | Ar | 403 | 0.601 | 0.0785 | Yes |
| 4 | Abca2 | 802 | 0.507 | 0.0814 | Yes |
| 5 | Bmp6 | 874 | 0.496 | 0.1005 | Yes |
| 6 | Gstk1 | 1047 | 0.470 | 0.1133 | Yes |
| 7 | Tfcp2l1 | 1546 | 0.406 | 0.1063 | Yes |
| 8 | Abca6 | 2028 | 0.359 | 0.0980 | Yes |
| 9 | Nedd4 | 2156 | 0.348 | 0.1075 | Yes |
| 10 | Lonp2 | 2203 | 0.343 | 0.1209 | Yes |
| 11 | Mlycd | 2683 | 0.308 | 0.1104 | Yes |
| 12 | Pipox | 2710 | 0.306 | 0.1232 | Yes |
| 13 | Pex7 | 2759 | 0.305 | 0.1348 | Yes |
| 14 | Cyp7b1 | 2924 | 0.294 | 0.1398 | Yes |
| 15 | Nr3c2 | 2996 | 0.289 | 0.1495 | Yes |
| 16 | Nr0b2 | 3005 | 0.288 | 0.1623 | Yes |
| 17 | Slc23a1 | 3014 | 0.288 | 0.1751 | Yes |
| 18 | Pex26 | 3109 | 0.282 | 0.1833 | Yes |
| 19 | Klf1 | 3182 | 0.278 | 0.1923 | Yes |
| 20 | Npc1 | 3267 | 0.273 | 0.2006 | Yes |
| 21 | Acsl1 | 3398 | 0.264 | 0.2060 | Yes |
| 22 | Idi1 | 3681 | 0.248 | 0.2029 | Yes |
| 23 | Lipe | 3855 | 0.238 | 0.2049 | Yes |
| 24 | Prdx5 | 4162 | 0.222 | 0.1994 | Yes |
| 25 | Fads1 | 4169 | 0.221 | 0.2093 | Yes |
| 26 | Gnpat | 4174 | 0.221 | 0.2192 | Yes |
| 27 | Sult2b1 | 4227 | 0.219 | 0.2266 | Yes |
| 28 | Abcd1 | 4429 | 0.208 | 0.2259 | No |
| 29 | Pex16 | 4889 | 0.186 | 0.2108 | No |
| 30 | Slc22a18 | 5427 | 0.160 | 0.1905 | No |
| 31 | Slc23a2 | 5502 | 0.157 | 0.1939 | No |
| 32 | Optn | 5522 | 0.156 | 0.2001 | No |
| 33 | Pfkm | 5599 | 0.152 | 0.2032 | No |
| 34 | Pnpla8 | 5653 | 0.150 | 0.2074 | No |
| 35 | Aldh9a1 | 5670 | 0.149 | 0.2134 | No |
| 36 | Ephx2 | 5820 | 0.142 | 0.2123 | No |
| 37 | Pecr | 5987 | 0.136 | 0.2100 | No |
| 38 | Lck | 6569 | 0.113 | 0.1852 | No |
| 39 | Aqp9 | 6579 | 0.112 | 0.1900 | No |
| 40 | Fdxr | 6643 | 0.110 | 0.1918 | No |
| 41 | Amacr | 7279 | 0.086 | 0.1631 | No |
| 42 | Hsd17b4 | 7287 | 0.086 | 0.1667 | No |
| 43 | Pex6 | 7603 | 0.074 | 0.1538 | No |
| 44 | Cat | 8398 | 0.044 | 0.1150 | No |
| 45 | Pex11g | 8493 | 0.041 | 0.1120 | No |
| 46 | Slc27a2 | 8538 | 0.040 | 0.1116 | No |
| 47 | Hacl1 | 8826 | 0.030 | 0.0982 | No |
| 48 | Dhcr24 | 8967 | 0.025 | 0.0921 | No |
| 49 | Isoc1 | 9086 | 0.021 | 0.0870 | No |
| 50 | Hsd17b11 | 9236 | 0.015 | 0.0800 | No |
| 51 | Rbp1 | 9983 | -0.011 | 0.0421 | No |
| 52 | Aldh1a1 | 10066 | -0.014 | 0.0385 | No |
| 53 | Crot | 10141 | -0.017 | 0.0355 | No |
| 54 | Abcd3 | 10261 | -0.019 | 0.0302 | No |
| 55 | Pxmp2 | 10370 | -0.024 | 0.0258 | No |
| 56 | Bcar3 | 10442 | -0.026 | 0.0233 | No |
| 57 | Nudt12 | 10486 | -0.028 | 0.0224 | No |
| 58 | Abcg4 | 10666 | -0.034 | 0.0147 | No |
| 59 | Pex11a | 10872 | -0.041 | 0.0060 | No |
| 60 | Acsl5 | 11019 | -0.046 | 0.0006 | No |
| 61 | Pex19 | 11790 | -0.073 | -0.0356 | No |
| 62 | Abca5 | 11936 | -0.079 | -0.0395 | No |
| 63 | Akr1d1 | 12089 | -0.084 | -0.0434 | No |
| 64 | Fads2 | 12229 | -0.090 | -0.0465 | No |
| 65 | Pex12 | 12368 | -0.096 | -0.0491 | No |
| 66 | Idh2 | 13419 | -0.139 | -0.0968 | No |
| 67 | Phyh | 14032 | -0.166 | -0.1207 | No |
| 68 | Atxn1 | 14046 | -0.167 | -0.1137 | No |
| 69 | Dio2 | 14568 | -0.191 | -0.1317 | No |
| 70 | Abca3 | 14689 | -0.196 | -0.1289 | No |
| 71 | Slc35b2 | 14835 | -0.203 | -0.1270 | No |
| 72 | Idh1 | 15246 | -0.223 | -0.1378 | No |
| 73 | Slc29a1 | 15617 | -0.246 | -0.1456 | No |
| 74 | Rxra | 15711 | -0.252 | -0.1388 | No |
| 75 | Gclm | 15735 | -0.254 | -0.1283 | No |
| 76 | Cyp39a1 | 15776 | -0.256 | -0.1185 | No |
| 77 | Sod1 | 15858 | -0.261 | -0.1107 | No |
| 78 | Abca9 | 15919 | -0.266 | -0.1015 | No |
| 79 | Slc27a5 | 16006 | -0.273 | -0.0934 | No |
| 80 | Nr1h4 | 16287 | -0.288 | -0.0946 | No |
| 81 | Paox | 16750 | -0.317 | -0.1038 | No |
| 82 | Scp2 | 17055 | -0.340 | -0.1038 | No |
| 83 | Retsat | 17058 | -0.340 | -0.0882 | No |
| 84 | Soat2 | 17303 | -0.360 | -0.0842 | No |
| 85 | Cyp27a1 | 17543 | -0.383 | -0.0789 | No |
| 86 | Efhc1 | 17597 | -0.388 | -0.0638 | No |
| 87 | Gnmt | 17729 | -0.398 | -0.0522 | No |
| 88 | Abca8b | 17879 | -0.414 | -0.0408 | No |
| 89 | Pex1 | 18175 | -0.449 | -0.0354 | No |
| 90 | Pex13 | 18288 | -0.465 | -0.0198 | No |
| 91 | Cyp46a1 | 18851 | -0.556 | -0.0231 | No |
| 92 | Abca1 | 19026 | -0.606 | -0.0042 | No |
| 93 | Abca4 | 19108 | -0.633 | 0.0208 | No |