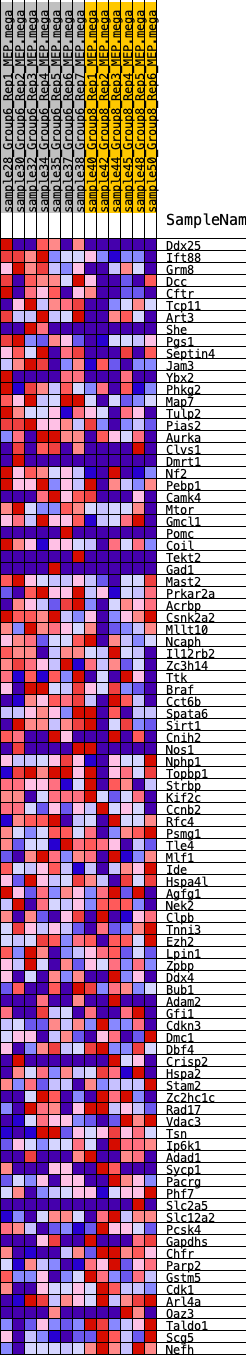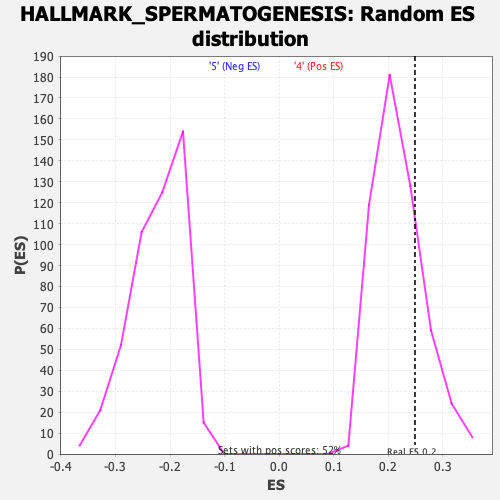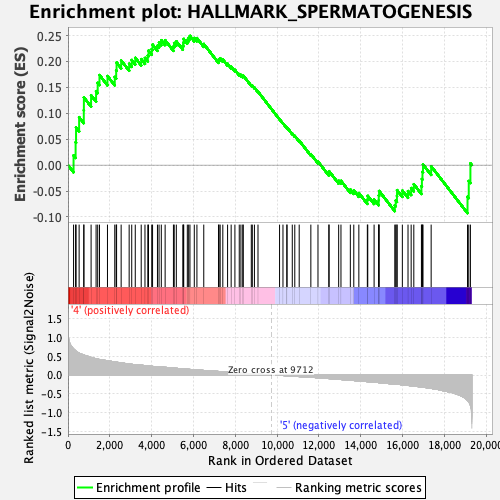
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.24938852 |
| Normalized Enrichment Score (NES) | 1.1333824 |
| Nominal p-value | 0.23709369 |
| FDR q-value | 0.7681681 |
| FWER p-Value | 0.973 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ddx25 | 266 | 0.692 | 0.0188 | Yes |
| 2 | Ift88 | 364 | 0.641 | 0.0441 | Yes |
| 3 | Grm8 | 385 | 0.631 | 0.0728 | Yes |
| 4 | Dcc | 532 | 0.576 | 0.0925 | Yes |
| 5 | Cftr | 755 | 0.529 | 0.1059 | Yes |
| 6 | Tcp11 | 760 | 0.527 | 0.1306 | Yes |
| 7 | Art3 | 1104 | 0.466 | 0.1348 | Yes |
| 8 | She | 1343 | 0.432 | 0.1428 | Yes |
| 9 | Pgs1 | 1420 | 0.422 | 0.1588 | Yes |
| 10 | Septin4 | 1505 | 0.410 | 0.1738 | Yes |
| 11 | Jam3 | 1887 | 0.380 | 0.1719 | Yes |
| 12 | Ybx2 | 2236 | 0.349 | 0.1703 | Yes |
| 13 | Phkg2 | 2303 | 0.341 | 0.1830 | Yes |
| 14 | Map7 | 2323 | 0.339 | 0.1980 | Yes |
| 15 | Tulp2 | 2541 | 0.323 | 0.2019 | Yes |
| 16 | Pias2 | 2924 | 0.292 | 0.1959 | Yes |
| 17 | Aurka | 3047 | 0.283 | 0.2029 | Yes |
| 18 | Clvs1 | 3216 | 0.274 | 0.2070 | Yes |
| 19 | Dmrt1 | 3500 | 0.258 | 0.2045 | Yes |
| 20 | Nf2 | 3680 | 0.248 | 0.2069 | Yes |
| 21 | Pebp1 | 3816 | 0.242 | 0.2113 | Yes |
| 22 | Camk4 | 3849 | 0.240 | 0.2210 | Yes |
| 23 | Mtor | 4004 | 0.229 | 0.2238 | Yes |
| 24 | Gmcl1 | 4043 | 0.227 | 0.2326 | Yes |
| 25 | Pomc | 4277 | 0.214 | 0.2305 | Yes |
| 26 | Coil | 4353 | 0.213 | 0.2367 | Yes |
| 27 | Tekt2 | 4461 | 0.208 | 0.2410 | Yes |
| 28 | Gad1 | 4644 | 0.202 | 0.2410 | Yes |
| 29 | Mast2 | 5041 | 0.182 | 0.2290 | Yes |
| 30 | Prkar2a | 5078 | 0.181 | 0.2357 | Yes |
| 31 | Acrbp | 5179 | 0.177 | 0.2388 | Yes |
| 32 | Csnk2a2 | 5490 | 0.163 | 0.2304 | Yes |
| 33 | Mllt10 | 5520 | 0.162 | 0.2365 | Yes |
| 34 | Ncaph | 5533 | 0.161 | 0.2435 | Yes |
| 35 | Il12rb2 | 5705 | 0.153 | 0.2418 | Yes |
| 36 | Zc3h14 | 5769 | 0.150 | 0.2456 | Yes |
| 37 | Ttk | 5831 | 0.147 | 0.2494 | Yes |
| 38 | Braf | 6038 | 0.140 | 0.2453 | No |
| 39 | Cct6b | 6164 | 0.134 | 0.2451 | No |
| 40 | Spata6 | 6491 | 0.120 | 0.2338 | No |
| 41 | Sirt1 | 7195 | 0.094 | 0.2016 | No |
| 42 | Cnih2 | 7222 | 0.092 | 0.2046 | No |
| 43 | Nos1 | 7275 | 0.090 | 0.2062 | No |
| 44 | Nphp1 | 7402 | 0.086 | 0.2037 | No |
| 45 | Topbp1 | 7630 | 0.077 | 0.1955 | No |
| 46 | Strbp | 7801 | 0.071 | 0.1900 | No |
| 47 | Kif2c | 7980 | 0.064 | 0.1838 | No |
| 48 | Ccnb2 | 8188 | 0.055 | 0.1756 | No |
| 49 | Rfc4 | 8267 | 0.052 | 0.1740 | No |
| 50 | Psmg1 | 8356 | 0.048 | 0.1717 | No |
| 51 | Tle4 | 8379 | 0.048 | 0.1728 | No |
| 52 | Mlf1 | 8764 | 0.034 | 0.1544 | No |
| 53 | Ide | 8812 | 0.031 | 0.1534 | No |
| 54 | Hspa4l | 8913 | 0.028 | 0.1495 | No |
| 55 | Agfg1 | 9084 | 0.023 | 0.1418 | No |
| 56 | Nek2 | 10115 | -0.007 | 0.0885 | No |
| 57 | Clpb | 10278 | -0.013 | 0.0807 | No |
| 58 | Tnni3 | 10466 | -0.020 | 0.0719 | No |
| 59 | Ezh2 | 10469 | -0.020 | 0.0727 | No |
| 60 | Lpin1 | 10723 | -0.029 | 0.0609 | No |
| 61 | Zpbp | 10843 | -0.033 | 0.0563 | No |
| 62 | Ddx4 | 11054 | -0.041 | 0.0473 | No |
| 63 | Bub1 | 11611 | -0.060 | 0.0212 | No |
| 64 | Adam2 | 11952 | -0.072 | 0.0069 | No |
| 65 | Gfi1 | 12472 | -0.094 | -0.0157 | No |
| 66 | Cdkn3 | 12482 | -0.094 | -0.0118 | No |
| 67 | Dmc1 | 12941 | -0.114 | -0.0302 | No |
| 68 | Dbf4 | 13051 | -0.119 | -0.0303 | No |
| 69 | Crisp2 | 13499 | -0.138 | -0.0470 | No |
| 70 | Hspa2 | 13664 | -0.146 | -0.0487 | No |
| 71 | Stam2 | 13906 | -0.157 | -0.0538 | No |
| 72 | Zc2hc1c | 14312 | -0.175 | -0.0666 | No |
| 73 | Rad17 | 14332 | -0.176 | -0.0593 | No |
| 74 | Vdac3 | 14635 | -0.189 | -0.0661 | No |
| 75 | Tsn | 14855 | -0.201 | -0.0680 | No |
| 76 | Ip6k1 | 14858 | -0.201 | -0.0586 | No |
| 77 | Adad1 | 14875 | -0.202 | -0.0499 | No |
| 78 | Sycp1 | 15625 | -0.240 | -0.0775 | No |
| 79 | Pacrg | 15669 | -0.243 | -0.0683 | No |
| 80 | Phf7 | 15727 | -0.246 | -0.0596 | No |
| 81 | Slc2a5 | 15733 | -0.246 | -0.0483 | No |
| 82 | Slc12a2 | 15986 | -0.260 | -0.0491 | No |
| 83 | Pcsk4 | 16257 | -0.277 | -0.0501 | No |
| 84 | Gapdhs | 16407 | -0.287 | -0.0442 | No |
| 85 | Chfr | 16534 | -0.296 | -0.0368 | No |
| 86 | Parp2 | 16898 | -0.319 | -0.0407 | No |
| 87 | Gstm5 | 16921 | -0.320 | -0.0267 | No |
| 88 | Cdk1 | 16948 | -0.323 | -0.0128 | No |
| 89 | Arl4a | 16973 | -0.324 | 0.0013 | No |
| 90 | Oaz3 | 17363 | -0.355 | -0.0022 | No |
| 91 | Taldo1 | 19102 | -0.672 | -0.0609 | No |
| 92 | Scg5 | 19161 | -0.706 | -0.0305 | No |
| 93 | Nefh | 19240 | -0.806 | 0.0035 | No |