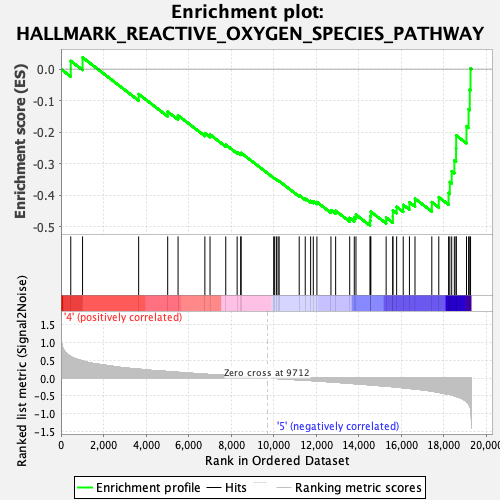
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.4964502 |
| Normalized Enrichment Score (NES) | -1.6122798 |
| Nominal p-value | 0.024390243 |
| FDR q-value | 0.19412142 |
| FWER p-Value | 0.198 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mpo | 459 | 0.602 | 0.0258 | No |
| 2 | Nqo1 | 1014 | 0.484 | 0.0370 | No |
| 3 | Sbno2 | 3652 | 0.249 | -0.0793 | No |
| 4 | Abcc1 | 5018 | 0.183 | -0.1351 | No |
| 5 | Msra | 5506 | 0.162 | -0.1470 | No |
| 6 | Ercc2 | 6767 | 0.109 | -0.2034 | No |
| 7 | Pfkp | 7010 | 0.101 | -0.2077 | No |
| 8 | Gsr | 7749 | 0.072 | -0.2400 | No |
| 9 | Mbp | 8285 | 0.051 | -0.2636 | No |
| 10 | Oxsr1 | 8452 | 0.045 | -0.2685 | No |
| 11 | Scaf4 | 8473 | 0.044 | -0.2659 | No |
| 12 | Gclm | 10004 | -0.003 | -0.3451 | No |
| 13 | G6pdx | 10049 | -0.005 | -0.3470 | No |
| 14 | Ipcef1 | 10132 | -0.007 | -0.3506 | No |
| 15 | Hmox2 | 10209 | -0.010 | -0.3537 | No |
| 16 | Gclc | 10260 | -0.012 | -0.3553 | No |
| 17 | Stk25 | 11204 | -0.045 | -0.4006 | No |
| 18 | Ndufs2 | 11488 | -0.056 | -0.4106 | No |
| 19 | Prnp | 11741 | -0.064 | -0.4184 | No |
| 20 | Srxn1 | 11867 | -0.069 | -0.4192 | No |
| 21 | Txnrd1 | 12037 | -0.075 | -0.4218 | No |
| 22 | Pdlim1 | 12697 | -0.104 | -0.4475 | No |
| 23 | Prdx4 | 12918 | -0.112 | -0.4496 | No |
| 24 | Selenos | 13579 | -0.141 | -0.4722 | No |
| 25 | Ptpa | 13794 | -0.152 | -0.4708 | No |
| 26 | Gpx4 | 13874 | -0.155 | -0.4621 | No |
| 27 | Cat | 14537 | -0.185 | -0.4812 | Yes |
| 28 | Txn1 | 14550 | -0.185 | -0.4665 | Yes |
| 29 | Glrx | 14574 | -0.186 | -0.4523 | Yes |
| 30 | Ftl1 | 15293 | -0.225 | -0.4711 | Yes |
| 31 | Prdx6 | 15607 | -0.238 | -0.4677 | Yes |
| 32 | Txnrd2 | 15609 | -0.239 | -0.4480 | Yes |
| 33 | Hhex | 15787 | -0.247 | -0.4368 | Yes |
| 34 | Fes | 16097 | -0.267 | -0.4309 | Yes |
| 35 | Prdx1 | 16389 | -0.286 | -0.4224 | Yes |
| 36 | Lsp1 | 16650 | -0.303 | -0.4108 | Yes |
| 37 | Mgst1 | 17441 | -0.362 | -0.4220 | Yes |
| 38 | Glrx2 | 17769 | -0.395 | -0.4063 | Yes |
| 39 | Ndufa6 | 18230 | -0.450 | -0.3931 | Yes |
| 40 | Sod2 | 18288 | -0.457 | -0.3583 | Yes |
| 41 | Egln2 | 18377 | -0.469 | -0.3241 | Yes |
| 42 | Cdkn2d | 18501 | -0.492 | -0.2899 | Yes |
| 43 | Junb | 18583 | -0.508 | -0.2522 | Yes |
| 44 | Lamtor5 | 18589 | -0.509 | -0.2104 | Yes |
| 45 | Sod1 | 19073 | -0.656 | -0.1813 | Yes |
| 46 | Prdx2 | 19171 | -0.716 | -0.1273 | Yes |
| 47 | Gpx3 | 19226 | -0.781 | -0.0656 | Yes |
| 48 | Atox1 | 19263 | -0.845 | 0.0023 | Yes |

