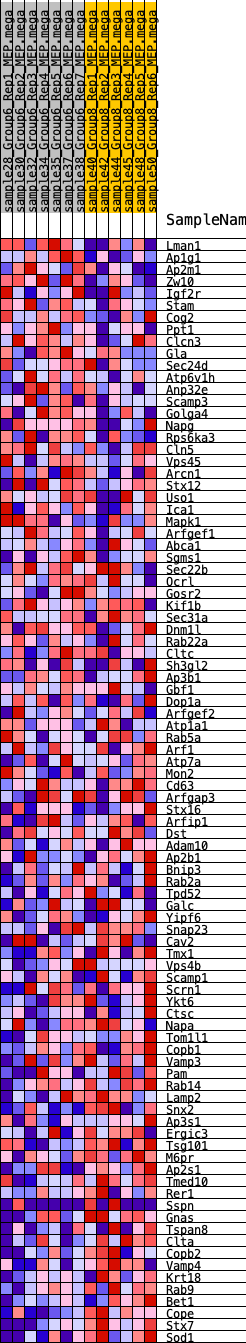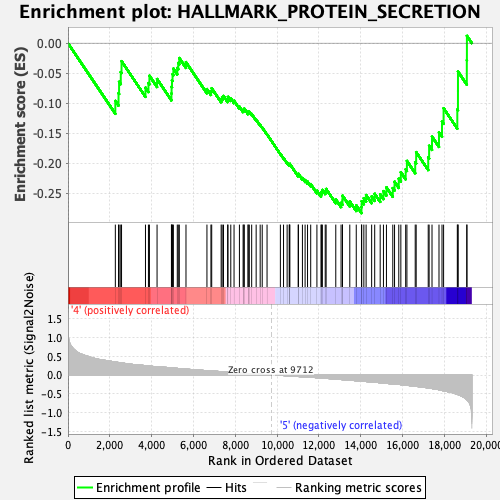
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.2833646 |
| Normalized Enrichment Score (NES) | -1.3121301 |
| Nominal p-value | 0.17171717 |
| FDR q-value | 0.3835519 |
| FWER p-Value | 0.767 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lman1 | 2263 | 0.346 | -0.0962 | No |
| 2 | Ap1g1 | 2416 | 0.332 | -0.0835 | No |
| 3 | Ap2m1 | 2443 | 0.330 | -0.0643 | No |
| 4 | Zw10 | 2524 | 0.324 | -0.0482 | No |
| 5 | Igf2r | 2560 | 0.321 | -0.0300 | No |
| 6 | Stam | 3704 | 0.246 | -0.0742 | No |
| 7 | Cog2 | 3846 | 0.240 | -0.0665 | No |
| 8 | Ppt1 | 3893 | 0.236 | -0.0542 | No |
| 9 | Clcn3 | 4258 | 0.215 | -0.0597 | No |
| 10 | Gla | 4945 | 0.187 | -0.0838 | No |
| 11 | Sec24d | 4953 | 0.187 | -0.0725 | No |
| 12 | Atp6v1h | 4972 | 0.186 | -0.0619 | No |
| 13 | Anp32e | 4999 | 0.184 | -0.0518 | No |
| 14 | Scamp3 | 5038 | 0.183 | -0.0424 | No |
| 15 | Golga4 | 5225 | 0.175 | -0.0411 | No |
| 16 | Napg | 5272 | 0.172 | -0.0328 | No |
| 17 | Rps6ka3 | 5322 | 0.170 | -0.0248 | No |
| 18 | Cln5 | 5641 | 0.156 | -0.0316 | No |
| 19 | Vps45 | 6643 | 0.114 | -0.0765 | No |
| 20 | Arcn1 | 6834 | 0.106 | -0.0798 | No |
| 21 | Stx12 | 6871 | 0.105 | -0.0751 | No |
| 22 | Uso1 | 7323 | 0.089 | -0.0930 | No |
| 23 | Ica1 | 7370 | 0.087 | -0.0900 | No |
| 24 | Mapk1 | 7434 | 0.085 | -0.0880 | No |
| 25 | Arfgef1 | 7634 | 0.077 | -0.0935 | No |
| 26 | Abca1 | 7648 | 0.077 | -0.0894 | No |
| 27 | Sgms1 | 7783 | 0.071 | -0.0919 | No |
| 28 | Sec22b | 7939 | 0.065 | -0.0960 | No |
| 29 | Ocrl | 8196 | 0.055 | -0.1059 | No |
| 30 | Gosr2 | 8372 | 0.048 | -0.1120 | No |
| 31 | Kif1b | 8408 | 0.046 | -0.1109 | No |
| 32 | Sec31a | 8426 | 0.046 | -0.1090 | No |
| 33 | Dnm1l | 8610 | 0.039 | -0.1161 | No |
| 34 | Rab22a | 8613 | 0.039 | -0.1138 | No |
| 35 | Cltc | 8669 | 0.037 | -0.1143 | No |
| 36 | Sh3gl2 | 8782 | 0.033 | -0.1181 | No |
| 37 | Ap3b1 | 8996 | 0.025 | -0.1276 | No |
| 38 | Gbf1 | 9188 | 0.018 | -0.1364 | No |
| 39 | Dop1a | 9286 | 0.015 | -0.1406 | No |
| 40 | Arfgef2 | 9517 | 0.007 | -0.1521 | No |
| 41 | Atp1a1 | 10154 | -0.008 | -0.1847 | No |
| 42 | Rab5a | 10305 | -0.014 | -0.1916 | No |
| 43 | Arf1 | 10477 | -0.020 | -0.1993 | No |
| 44 | Atp7a | 10576 | -0.024 | -0.2029 | No |
| 45 | Mon2 | 10596 | -0.025 | -0.2023 | No |
| 46 | Cd63 | 10604 | -0.025 | -0.2012 | No |
| 47 | Arfgap3 | 11006 | -0.039 | -0.2196 | No |
| 48 | Stx16 | 11018 | -0.039 | -0.2178 | No |
| 49 | Arfip1 | 11208 | -0.045 | -0.2248 | No |
| 50 | Dst | 11336 | -0.050 | -0.2283 | No |
| 51 | Adam10 | 11453 | -0.055 | -0.2309 | No |
| 52 | Ap2b1 | 11604 | -0.059 | -0.2350 | No |
| 53 | Bnip3 | 11901 | -0.071 | -0.2460 | No |
| 54 | Rab2a | 12096 | -0.078 | -0.2512 | No |
| 55 | Tpd52 | 12124 | -0.079 | -0.2477 | No |
| 56 | Galc | 12162 | -0.080 | -0.2446 | No |
| 57 | Yipf6 | 12298 | -0.086 | -0.2463 | No |
| 58 | Snap23 | 12341 | -0.088 | -0.2430 | No |
| 59 | Cav2 | 12803 | -0.108 | -0.2603 | No |
| 60 | Tmx1 | 13056 | -0.119 | -0.2660 | No |
| 61 | Vps4b | 13123 | -0.122 | -0.2618 | No |
| 62 | Scamp1 | 13124 | -0.122 | -0.2542 | No |
| 63 | Scrn1 | 13473 | -0.137 | -0.2638 | No |
| 64 | Ykt6 | 13782 | -0.152 | -0.2704 | No |
| 65 | Ctsc | 14032 | -0.163 | -0.2732 | Yes |
| 66 | Napa | 14043 | -0.163 | -0.2636 | Yes |
| 67 | Tom1l1 | 14145 | -0.168 | -0.2584 | Yes |
| 68 | Copb1 | 14253 | -0.172 | -0.2532 | Yes |
| 69 | Vamp3 | 14518 | -0.184 | -0.2555 | Yes |
| 70 | Pam | 14664 | -0.191 | -0.2511 | Yes |
| 71 | Rab14 | 14926 | -0.205 | -0.2519 | Yes |
| 72 | Lamp2 | 15082 | -0.213 | -0.2467 | Yes |
| 73 | Snx2 | 15225 | -0.221 | -0.2403 | Yes |
| 74 | Ap3s1 | 15529 | -0.234 | -0.2415 | Yes |
| 75 | Ergic3 | 15613 | -0.239 | -0.2310 | Yes |
| 76 | Tsg101 | 15815 | -0.248 | -0.2259 | Yes |
| 77 | M6pr | 15914 | -0.255 | -0.2151 | Yes |
| 78 | Ap2s1 | 16145 | -0.270 | -0.2103 | Yes |
| 79 | Tmed10 | 16204 | -0.274 | -0.1963 | Yes |
| 80 | Rer1 | 16601 | -0.299 | -0.1982 | Yes |
| 81 | Sspn | 16647 | -0.303 | -0.1817 | Yes |
| 82 | Gnas | 17223 | -0.342 | -0.1903 | Yes |
| 83 | Tspan8 | 17266 | -0.346 | -0.1709 | Yes |
| 84 | Clta | 17404 | -0.358 | -0.1557 | Yes |
| 85 | Copb2 | 17738 | -0.391 | -0.1486 | Yes |
| 86 | Vamp4 | 17882 | -0.409 | -0.1306 | Yes |
| 87 | Krt18 | 17958 | -0.417 | -0.1085 | Yes |
| 88 | Rab9 | 18614 | -0.515 | -0.1105 | Yes |
| 89 | Bet1 | 18645 | -0.520 | -0.0796 | Yes |
| 90 | Cope | 18646 | -0.520 | -0.0473 | Yes |
| 91 | Stx7 | 19068 | -0.652 | -0.0285 | Yes |
| 92 | Sod1 | 19073 | -0.656 | 0.0122 | Yes |