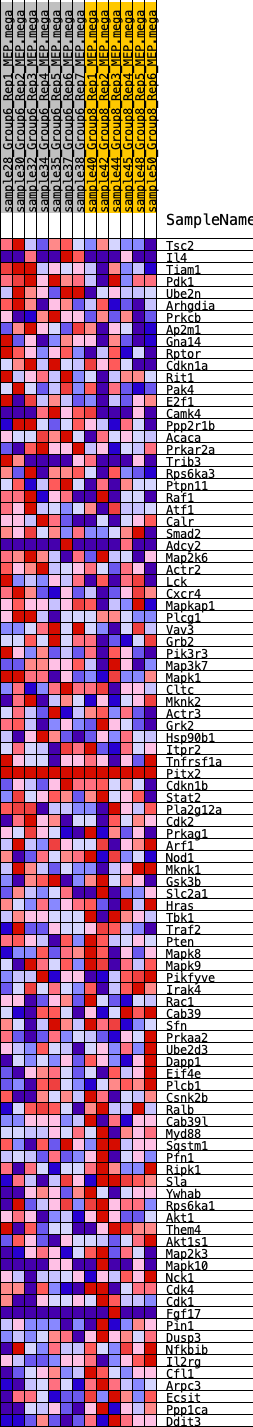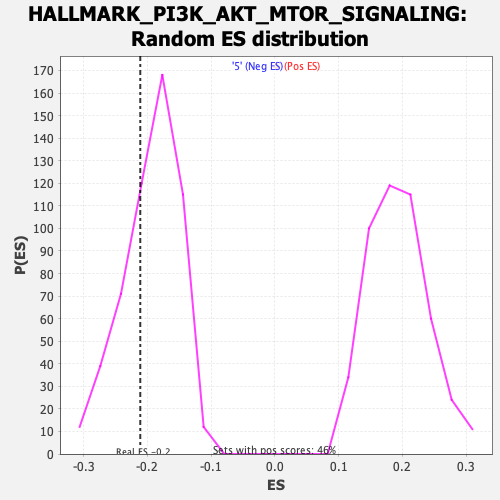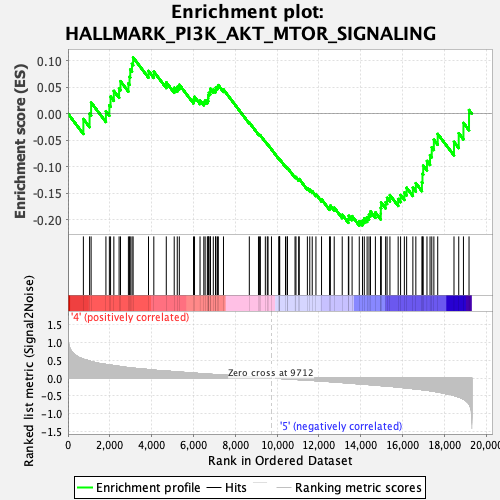
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.21113762 |
| Normalized Enrichment Score (NES) | -1.0894659 |
| Nominal p-value | 0.30167598 |
| FDR q-value | 0.5004098 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tsc2 | 735 | 0.532 | -0.0100 | No |
| 2 | Il4 | 1032 | 0.482 | 0.0001 | No |
| 3 | Tiam1 | 1105 | 0.466 | 0.0211 | No |
| 4 | Pdk1 | 1813 | 0.383 | 0.0047 | No |
| 5 | Ube2n | 1978 | 0.372 | 0.0159 | No |
| 6 | Arhgdia | 2035 | 0.364 | 0.0323 | No |
| 7 | Prkcb | 2192 | 0.353 | 0.0430 | No |
| 8 | Ap2m1 | 2443 | 0.330 | 0.0475 | No |
| 9 | Gna14 | 2510 | 0.325 | 0.0613 | No |
| 10 | Rptor | 2888 | 0.294 | 0.0572 | No |
| 11 | Cdkn1a | 2943 | 0.290 | 0.0698 | No |
| 12 | Rit1 | 2976 | 0.288 | 0.0835 | No |
| 13 | Pak4 | 3059 | 0.282 | 0.0941 | No |
| 14 | E2f1 | 3113 | 0.278 | 0.1062 | No |
| 15 | Camk4 | 3849 | 0.240 | 0.0806 | No |
| 16 | Ppp2r1b | 4103 | 0.224 | 0.0794 | No |
| 17 | Acaca | 4699 | 0.201 | 0.0591 | No |
| 18 | Prkar2a | 5078 | 0.181 | 0.0490 | No |
| 19 | Trib3 | 5224 | 0.175 | 0.0507 | No |
| 20 | Rps6ka3 | 5322 | 0.170 | 0.0547 | No |
| 21 | Ptpn11 | 6001 | 0.141 | 0.0269 | No |
| 22 | Raf1 | 6049 | 0.139 | 0.0318 | No |
| 23 | Atf1 | 6313 | 0.128 | 0.0249 | No |
| 24 | Calr | 6494 | 0.120 | 0.0219 | No |
| 25 | Smad2 | 6556 | 0.117 | 0.0249 | No |
| 26 | Adcy2 | 6663 | 0.114 | 0.0254 | No |
| 27 | Map2k6 | 6703 | 0.112 | 0.0294 | No |
| 28 | Actr2 | 6706 | 0.112 | 0.0352 | No |
| 29 | Lck | 6738 | 0.111 | 0.0394 | No |
| 30 | Cxcr4 | 6803 | 0.107 | 0.0418 | No |
| 31 | Mapkap1 | 6809 | 0.107 | 0.0472 | No |
| 32 | Plcg1 | 6947 | 0.103 | 0.0456 | No |
| 33 | Vav3 | 7052 | 0.099 | 0.0454 | No |
| 34 | Grb2 | 7072 | 0.098 | 0.0496 | No |
| 35 | Pik3r3 | 7145 | 0.096 | 0.0510 | No |
| 36 | Map3k7 | 7187 | 0.094 | 0.0538 | No |
| 37 | Mapk1 | 7434 | 0.085 | 0.0455 | No |
| 38 | Cltc | 8669 | 0.037 | -0.0167 | No |
| 39 | Mknk2 | 9107 | 0.021 | -0.0384 | No |
| 40 | Actr3 | 9160 | 0.019 | -0.0400 | No |
| 41 | Grk2 | 9198 | 0.018 | -0.0410 | No |
| 42 | Hsp90b1 | 9435 | 0.010 | -0.0528 | No |
| 43 | Itpr2 | 9529 | 0.006 | -0.0573 | No |
| 44 | Tnfrsf1a | 9564 | 0.005 | -0.0588 | No |
| 45 | Pitx2 | 9729 | 0.000 | -0.0673 | No |
| 46 | Cdkn1b | 10078 | -0.006 | -0.0851 | No |
| 47 | Stat2 | 10100 | -0.007 | -0.0859 | No |
| 48 | Pla2g12a | 10110 | -0.007 | -0.0860 | No |
| 49 | Cdk2 | 10123 | -0.007 | -0.0862 | No |
| 50 | Prkag1 | 10399 | -0.018 | -0.0996 | No |
| 51 | Arf1 | 10477 | -0.020 | -0.1025 | No |
| 52 | Nod1 | 10492 | -0.021 | -0.1021 | No |
| 53 | Mknk1 | 10855 | -0.033 | -0.1192 | No |
| 54 | Gsk3b | 10899 | -0.035 | -0.1196 | No |
| 55 | Slc2a1 | 11030 | -0.040 | -0.1242 | No |
| 56 | Hras | 11055 | -0.041 | -0.1233 | No |
| 57 | Tbk1 | 11445 | -0.054 | -0.1407 | No |
| 58 | Traf2 | 11559 | -0.058 | -0.1435 | No |
| 59 | Pten | 11677 | -0.062 | -0.1463 | No |
| 60 | Mapk8 | 11854 | -0.069 | -0.1518 | No |
| 61 | Mapk9 | 12122 | -0.079 | -0.1615 | No |
| 62 | Pikfyve | 12505 | -0.095 | -0.1764 | No |
| 63 | Irak4 | 12545 | -0.096 | -0.1733 | No |
| 64 | Rac1 | 12728 | -0.105 | -0.1772 | No |
| 65 | Cab39 | 13111 | -0.121 | -0.1906 | No |
| 66 | Sfn | 13405 | -0.133 | -0.1988 | No |
| 67 | Prkaa2 | 13425 | -0.135 | -0.1927 | No |
| 68 | Ube2d3 | 13583 | -0.141 | -0.1933 | No |
| 69 | Dapp1 | 13926 | -0.158 | -0.2028 | Yes |
| 70 | Eif4e | 14080 | -0.165 | -0.2020 | Yes |
| 71 | Plcb1 | 14166 | -0.168 | -0.1974 | Yes |
| 72 | Csnk2b | 14301 | -0.175 | -0.1951 | Yes |
| 73 | Ralb | 14392 | -0.178 | -0.1904 | Yes |
| 74 | Cab39l | 14462 | -0.181 | -0.1843 | Yes |
| 75 | Myd88 | 14697 | -0.193 | -0.1863 | Yes |
| 76 | Sqstm1 | 14954 | -0.206 | -0.1887 | Yes |
| 77 | Pfn1 | 14956 | -0.206 | -0.1778 | Yes |
| 78 | Ripk1 | 14970 | -0.207 | -0.1675 | Yes |
| 79 | Sla | 15187 | -0.219 | -0.1671 | Yes |
| 80 | Ywhab | 15255 | -0.223 | -0.1588 | Yes |
| 81 | Rps6ka1 | 15392 | -0.230 | -0.1536 | Yes |
| 82 | Akt1 | 15786 | -0.247 | -0.1610 | Yes |
| 83 | Them4 | 15901 | -0.254 | -0.1534 | Yes |
| 84 | Akt1s1 | 16082 | -0.266 | -0.1487 | Yes |
| 85 | Map2k3 | 16191 | -0.273 | -0.1398 | Yes |
| 86 | Mapk10 | 16489 | -0.293 | -0.1397 | Yes |
| 87 | Nck1 | 16631 | -0.302 | -0.1311 | Yes |
| 88 | Cdk4 | 16922 | -0.321 | -0.1291 | Yes |
| 89 | Cdk1 | 16948 | -0.323 | -0.1133 | Yes |
| 90 | Fgf17 | 16983 | -0.325 | -0.0978 | Yes |
| 91 | Pin1 | 17163 | -0.338 | -0.0892 | Yes |
| 92 | Dusp3 | 17310 | -0.350 | -0.0782 | Yes |
| 93 | Nfkbib | 17393 | -0.358 | -0.0634 | Yes |
| 94 | Il2rg | 17493 | -0.367 | -0.0491 | Yes |
| 95 | Cfl1 | 17677 | -0.384 | -0.0382 | Yes |
| 96 | Arpc3 | 18455 | -0.482 | -0.0531 | Yes |
| 97 | Ecsit | 18682 | -0.526 | -0.0369 | Yes |
| 98 | Ppp1ca | 18907 | -0.586 | -0.0175 | Yes |
| 99 | Ddit3 | 19174 | -0.720 | 0.0069 | Yes |