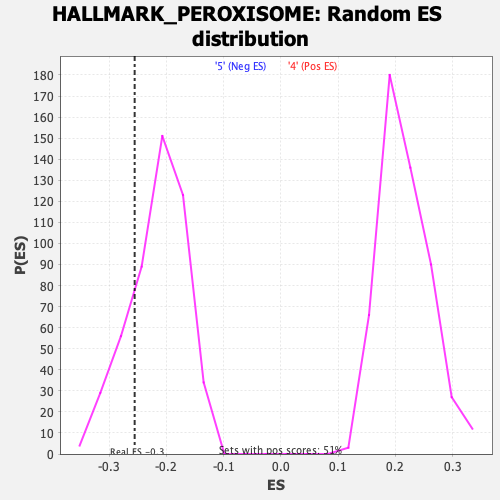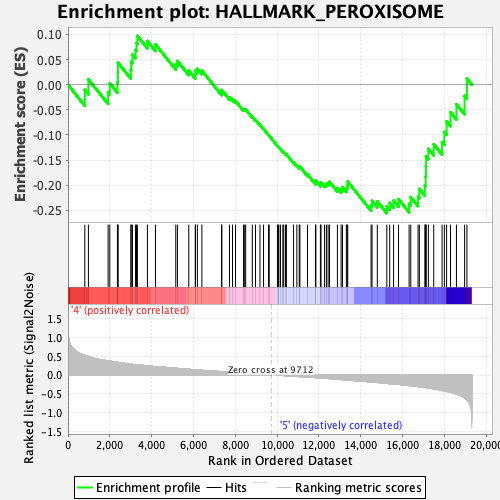
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.255356 |
| Normalized Enrichment Score (NES) | -1.1857492 |
| Nominal p-value | 0.2037037 |
| FDR q-value | 0.3910934 |
| FWER p-Value | 0.946 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acsl1 | 804 | 0.520 | -0.0102 | No |
| 2 | Abcd2 | 976 | 0.489 | 0.0106 | No |
| 3 | Itgb1bp1 | 1918 | 0.377 | -0.0153 | No |
| 4 | Pex5 | 1999 | 0.369 | 0.0030 | No |
| 5 | Dhcr24 | 2363 | 0.336 | 0.0045 | No |
| 6 | Slc25a17 | 2386 | 0.334 | 0.0237 | No |
| 7 | Abcb9 | 2389 | 0.334 | 0.0439 | No |
| 8 | Slc25a19 | 3005 | 0.286 | 0.0293 | No |
| 9 | Fdps | 3022 | 0.284 | 0.0458 | No |
| 10 | Slc23a2 | 3082 | 0.280 | 0.0597 | No |
| 11 | Hsd11b2 | 3226 | 0.273 | 0.0689 | No |
| 12 | Abcc5 | 3275 | 0.270 | 0.0829 | No |
| 13 | Idh1 | 3318 | 0.267 | 0.0970 | No |
| 14 | Smarcc1 | 3797 | 0.244 | 0.0869 | No |
| 15 | Sema3c | 4181 | 0.219 | 0.0804 | No |
| 16 | Fads1 | 5153 | 0.178 | 0.0407 | No |
| 17 | Abcd1 | 5232 | 0.174 | 0.0472 | No |
| 18 | Gstk1 | 5775 | 0.150 | 0.0281 | No |
| 19 | Idi1 | 6086 | 0.138 | 0.0204 | No |
| 20 | Elovl5 | 6094 | 0.137 | 0.0283 | No |
| 21 | Mlycd | 6186 | 0.133 | 0.0317 | No |
| 22 | Abcd3 | 6402 | 0.124 | 0.0281 | No |
| 23 | Retsat | 7347 | 0.088 | -0.0157 | No |
| 24 | Acaa1a | 7352 | 0.088 | -0.0105 | No |
| 25 | Pex14 | 7722 | 0.073 | -0.0253 | No |
| 26 | Fis1 | 7865 | 0.068 | -0.0285 | No |
| 27 | Crat | 8009 | 0.062 | -0.0321 | No |
| 28 | Siah1a | 8392 | 0.047 | -0.0491 | No |
| 29 | Esr2 | 8448 | 0.045 | -0.0493 | No |
| 30 | Ywhah | 8496 | 0.043 | -0.0491 | No |
| 31 | Ide | 8812 | 0.031 | -0.0636 | No |
| 32 | Atxn1 | 8964 | 0.026 | -0.0698 | No |
| 33 | Cln8 | 9179 | 0.019 | -0.0798 | No |
| 34 | Hsd3b7 | 9349 | 0.012 | -0.0879 | No |
| 35 | Acsl4 | 9590 | 0.004 | -0.1001 | No |
| 36 | Dlg4 | 9620 | 0.003 | -0.1014 | No |
| 37 | Top2a | 10014 | -0.004 | -0.1217 | No |
| 38 | Msh2 | 10060 | -0.005 | -0.1237 | No |
| 39 | Acot8 | 10081 | -0.006 | -0.1244 | No |
| 40 | Acsl5 | 10158 | -0.008 | -0.1278 | No |
| 41 | Sult2b1 | 10273 | -0.013 | -0.1330 | No |
| 42 | Ctbp1 | 10314 | -0.014 | -0.1342 | No |
| 43 | Hsd17b4 | 10397 | -0.018 | -0.1374 | No |
| 44 | Acox1 | 10442 | -0.019 | -0.1385 | No |
| 45 | Abcb1a | 10784 | -0.031 | -0.1544 | No |
| 46 | Ephx2 | 10941 | -0.037 | -0.1603 | No |
| 47 | Hras | 11055 | -0.041 | -0.1637 | No |
| 48 | Ech1 | 11088 | -0.042 | -0.1628 | No |
| 49 | Ercc1 | 11452 | -0.055 | -0.1783 | No |
| 50 | Rdh11 | 11831 | -0.068 | -0.1938 | No |
| 51 | Ehhadh | 11852 | -0.069 | -0.1907 | No |
| 52 | Eci2 | 12067 | -0.077 | -0.1972 | No |
| 53 | Pex6 | 12097 | -0.078 | -0.1940 | No |
| 54 | Pex13 | 12268 | -0.085 | -0.1976 | No |
| 55 | Cln6 | 12355 | -0.089 | -0.1967 | No |
| 56 | Gnpat | 12436 | -0.092 | -0.1953 | No |
| 57 | Pex2 | 12502 | -0.095 | -0.1929 | No |
| 58 | Pabpc1 | 12881 | -0.111 | -0.2058 | No |
| 59 | Lonp2 | 13058 | -0.119 | -0.2077 | No |
| 60 | Vps4b | 13123 | -0.122 | -0.2037 | No |
| 61 | Isoc1 | 13309 | -0.129 | -0.2054 | No |
| 62 | Cdk7 | 13348 | -0.131 | -0.1994 | No |
| 63 | Aldh9a1 | 13375 | -0.132 | -0.1928 | No |
| 64 | Idh2 | 14491 | -0.183 | -0.2396 | No |
| 65 | Cat | 14537 | -0.185 | -0.2307 | No |
| 66 | Ercc3 | 14789 | -0.197 | -0.2318 | No |
| 67 | Prdx5 | 15243 | -0.222 | -0.2419 | Yes |
| 68 | Tspo | 15381 | -0.230 | -0.2350 | Yes |
| 69 | Abcb4 | 15566 | -0.235 | -0.2303 | Yes |
| 70 | Dhrs3 | 15803 | -0.248 | -0.2275 | Yes |
| 71 | Slc35b2 | 16309 | -0.280 | -0.2367 | Yes |
| 72 | Prdx1 | 16389 | -0.286 | -0.2234 | Yes |
| 73 | Mvp | 16737 | -0.307 | -0.2228 | Yes |
| 74 | Cnbp | 16798 | -0.311 | -0.2070 | Yes |
| 75 | Hsd17b11 | 17062 | -0.330 | -0.2006 | Yes |
| 76 | Cacna1b | 17099 | -0.332 | -0.1822 | Yes |
| 77 | Pex11a | 17113 | -0.334 | -0.1626 | Yes |
| 78 | Aldh1a1 | 17120 | -0.334 | -0.1426 | Yes |
| 79 | Cadm1 | 17230 | -0.343 | -0.1274 | Yes |
| 80 | Hmgcl | 17487 | -0.367 | -0.1184 | Yes |
| 81 | Bcl10 | 17885 | -0.410 | -0.1141 | Yes |
| 82 | Slc25a4 | 17994 | -0.423 | -0.0940 | Yes |
| 83 | Pex11b | 18102 | -0.437 | -0.0729 | Yes |
| 84 | Sod2 | 18288 | -0.457 | -0.0547 | Yes |
| 85 | Scp2 | 18572 | -0.506 | -0.0387 | Yes |
| 86 | Nudt19 | 18963 | -0.606 | -0.0221 | Yes |
| 87 | Sod1 | 19073 | -0.656 | 0.0122 | Yes |