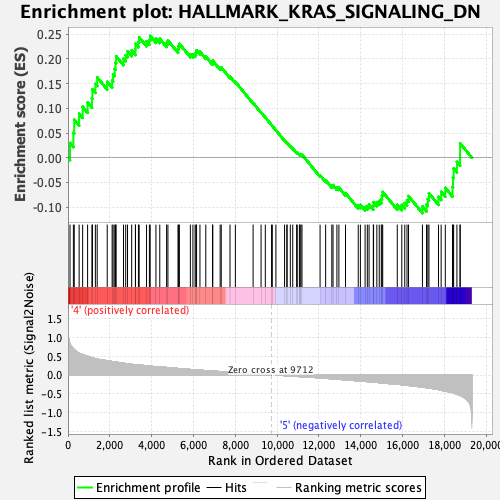
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.24682687 |
| Normalized Enrichment Score (NES) | 1.0438511 |
| Nominal p-value | 0.35091278 |
| FDR q-value | 0.64218867 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Efhd1 | 96 | 0.828 | 0.0298 | Yes |
| 2 | Tent5c | 257 | 0.699 | 0.0508 | Yes |
| 3 | Slc29a3 | 298 | 0.675 | 0.0771 | Yes |
| 4 | Dcc | 532 | 0.576 | 0.0892 | Yes |
| 5 | Dtnb | 700 | 0.541 | 0.1033 | Yes |
| 6 | Tgm1 | 937 | 0.498 | 0.1119 | Yes |
| 7 | Zfp112 | 1145 | 0.460 | 0.1205 | Yes |
| 8 | Myo15a | 1168 | 0.455 | 0.1384 | Yes |
| 9 | Macroh2a2 | 1308 | 0.435 | 0.1495 | Yes |
| 10 | Coq8a | 1395 | 0.425 | 0.1629 | Yes |
| 11 | Plag1 | 1875 | 0.381 | 0.1540 | Yes |
| 12 | Lgals7 | 2110 | 0.358 | 0.1569 | Yes |
| 13 | Sphk2 | 2157 | 0.354 | 0.1694 | Yes |
| 14 | Ybx2 | 2236 | 0.349 | 0.1800 | Yes |
| 15 | Mefv | 2272 | 0.345 | 0.1927 | Yes |
| 16 | Cd80 | 2301 | 0.342 | 0.2056 | Yes |
| 17 | Kmt2d | 2654 | 0.312 | 0.2004 | Yes |
| 18 | Bard1 | 2759 | 0.304 | 0.2077 | Yes |
| 19 | Entpd7 | 2846 | 0.297 | 0.2157 | Yes |
| 20 | Tcf7l1 | 3045 | 0.283 | 0.2173 | Yes |
| 21 | Ngb | 3224 | 0.273 | 0.2195 | Yes |
| 22 | Hsd11b2 | 3226 | 0.273 | 0.2310 | Yes |
| 23 | Camk1d | 3368 | 0.263 | 0.2347 | Yes |
| 24 | Slc25a23 | 3402 | 0.262 | 0.2440 | Yes |
| 25 | Sptbn2 | 3755 | 0.246 | 0.2360 | Yes |
| 26 | Arpp21 | 3892 | 0.236 | 0.2389 | Yes |
| 27 | Idua | 3929 | 0.234 | 0.2468 | Yes |
| 28 | Gpr19 | 4204 | 0.218 | 0.2417 | No |
| 29 | Celsr2 | 4388 | 0.211 | 0.2411 | No |
| 30 | Slc12a3 | 4714 | 0.200 | 0.2326 | No |
| 31 | Kcnn1 | 4777 | 0.196 | 0.2376 | No |
| 32 | Stag3 | 5262 | 0.173 | 0.2197 | No |
| 33 | Ptprj | 5278 | 0.172 | 0.2261 | No |
| 34 | Zbtb16 | 5325 | 0.170 | 0.2309 | No |
| 35 | Clstn3 | 5850 | 0.147 | 0.2097 | No |
| 36 | Tex15 | 5973 | 0.142 | 0.2094 | No |
| 37 | Tenm2 | 6076 | 0.138 | 0.2099 | No |
| 38 | Lfng | 6119 | 0.136 | 0.2134 | No |
| 39 | Slc6a3 | 6147 | 0.135 | 0.2177 | No |
| 40 | Bmpr1b | 6309 | 0.128 | 0.2147 | No |
| 41 | Cdkal1 | 6588 | 0.116 | 0.2051 | No |
| 42 | Btg2 | 6912 | 0.104 | 0.1926 | No |
| 43 | Msh5 | 6921 | 0.104 | 0.1966 | No |
| 44 | Nos1 | 7275 | 0.090 | 0.1820 | No |
| 45 | Fggy | 7327 | 0.089 | 0.1831 | No |
| 46 | Htr1b | 7746 | 0.072 | 0.1643 | No |
| 47 | Rsad2 | 8002 | 0.063 | 0.1537 | No |
| 48 | Mfsd6 | 8850 | 0.030 | 0.1109 | No |
| 49 | Thrb | 9231 | 0.017 | 0.0918 | No |
| 50 | Cyp39a1 | 9434 | 0.010 | 0.0817 | No |
| 51 | Capn9 | 9733 | 0.000 | 0.0662 | No |
| 52 | Htr1d | 9768 | 0.000 | 0.0644 | No |
| 53 | Mast3 | 9940 | -0.001 | 0.0555 | No |
| 54 | Atp4a | 10354 | -0.016 | 0.0347 | No |
| 55 | Tnni3 | 10466 | -0.020 | 0.0298 | No |
| 56 | Tfcp2l1 | 10472 | -0.020 | 0.0303 | No |
| 57 | Asb7 | 10630 | -0.026 | 0.0232 | No |
| 58 | Col2a1 | 10745 | -0.030 | 0.0186 | No |
| 59 | Serpinb2 | 10932 | -0.036 | 0.0104 | No |
| 60 | Pde6b | 10960 | -0.037 | 0.0106 | No |
| 61 | Mthfr | 11063 | -0.041 | 0.0070 | No |
| 62 | Nrip2 | 11093 | -0.043 | 0.0073 | No |
| 63 | P2rx6 | 11135 | -0.044 | 0.0070 | No |
| 64 | Ccdc106 | 11187 | -0.045 | 0.0062 | No |
| 65 | Prodh | 12052 | -0.076 | -0.0356 | No |
| 66 | Edar | 12318 | -0.087 | -0.0457 | No |
| 67 | Itgb1bp2 | 12613 | -0.099 | -0.0569 | No |
| 68 | Brdt | 12671 | -0.102 | -0.0555 | No |
| 69 | Vps50 | 12857 | -0.110 | -0.0605 | No |
| 70 | Gp1ba | 12952 | -0.114 | -0.0606 | No |
| 71 | Tg | 13268 | -0.127 | -0.0717 | No |
| 72 | Fgfr3 | 13877 | -0.155 | -0.0968 | No |
| 73 | Synpo | 13980 | -0.161 | -0.0954 | No |
| 74 | Pdk2 | 14205 | -0.170 | -0.0999 | No |
| 75 | Zc2hc1c | 14312 | -0.175 | -0.0980 | No |
| 76 | Kcnd1 | 14394 | -0.178 | -0.0947 | No |
| 77 | Epha5 | 14598 | -0.188 | -0.0974 | No |
| 78 | Abcg4 | 14610 | -0.188 | -0.0901 | No |
| 79 | Cpeb3 | 14766 | -0.196 | -0.0899 | No |
| 80 | Egf | 14881 | -0.202 | -0.0873 | No |
| 81 | Nr4a2 | 14979 | -0.208 | -0.0837 | No |
| 82 | Nr6a1 | 15012 | -0.210 | -0.0765 | No |
| 83 | Sidt1 | 15047 | -0.212 | -0.0694 | No |
| 84 | Cpa2 | 15743 | -0.246 | -0.0952 | No |
| 85 | Gamt | 15960 | -0.258 | -0.0956 | No |
| 86 | Ypel1 | 16102 | -0.267 | -0.0917 | No |
| 87 | Skil | 16209 | -0.274 | -0.0858 | No |
| 88 | Prkn | 16281 | -0.277 | -0.0778 | No |
| 89 | Snn | 16946 | -0.322 | -0.0988 | No |
| 90 | Copz2 | 17142 | -0.336 | -0.0948 | No |
| 91 | Klk8 | 17206 | -0.341 | -0.0838 | No |
| 92 | Gtf3c5 | 17258 | -0.346 | -0.0719 | No |
| 93 | Thnsl2 | 17714 | -0.388 | -0.0793 | No |
| 94 | Sgk1 | 17841 | -0.404 | -0.0688 | No |
| 95 | Mx2 | 18039 | -0.431 | -0.0610 | No |
| 96 | Selenop | 18383 | -0.471 | -0.0591 | No |
| 97 | Grid2 | 18408 | -0.475 | -0.0404 | No |
| 98 | Tgfb2 | 18438 | -0.479 | -0.0217 | No |
| 99 | Ryr1 | 18595 | -0.511 | -0.0084 | No |
| 100 | Slc16a7 | 18747 | -0.539 | 0.0064 | No |
| 101 | Ryr2 | 18748 | -0.539 | 0.0291 | No |

