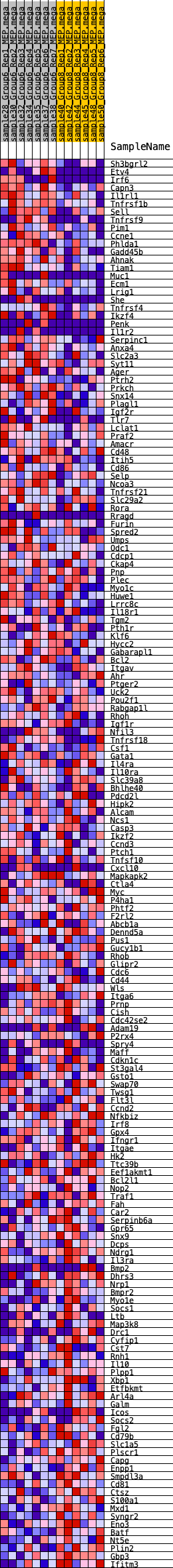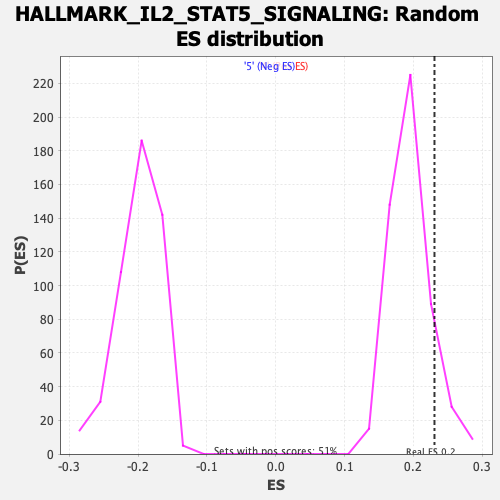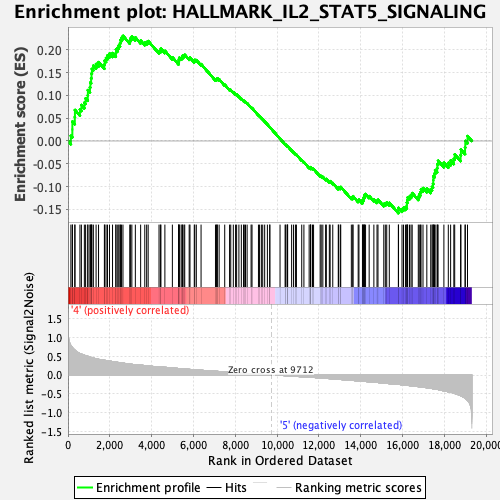
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | 0.23052235 |
| Normalized Enrichment Score (NES) | 1.1771452 |
| Nominal p-value | 0.11089494 |
| FDR q-value | 0.76538455 |
| FWER p-Value | 0.944 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sh3bgrl2 | 138 | 0.777 | 0.0115 | Yes |
| 2 | Etv4 | 208 | 0.725 | 0.0253 | Yes |
| 3 | Irf6 | 209 | 0.725 | 0.0427 | Yes |
| 4 | Capn3 | 326 | 0.659 | 0.0525 | Yes |
| 5 | Il1rl1 | 332 | 0.656 | 0.0680 | Yes |
| 6 | Tnfrsf1b | 571 | 0.566 | 0.0692 | Yes |
| 7 | Sell | 640 | 0.554 | 0.0789 | Yes |
| 8 | Tnfrsf9 | 782 | 0.523 | 0.0841 | Yes |
| 9 | Pim1 | 842 | 0.516 | 0.0935 | Yes |
| 10 | Ccne1 | 945 | 0.496 | 0.1000 | Yes |
| 11 | Phlda1 | 951 | 0.495 | 0.1117 | Yes |
| 12 | Gadd45b | 1050 | 0.476 | 0.1180 | Yes |
| 13 | Ahnak | 1069 | 0.473 | 0.1284 | Yes |
| 14 | Tiam1 | 1105 | 0.466 | 0.1378 | Yes |
| 15 | Muc1 | 1124 | 0.463 | 0.1480 | Yes |
| 16 | Ecm1 | 1143 | 0.460 | 0.1581 | Yes |
| 17 | Lrig1 | 1212 | 0.448 | 0.1653 | Yes |
| 18 | She | 1343 | 0.432 | 0.1689 | Yes |
| 19 | Tnfrsf4 | 1460 | 0.417 | 0.1729 | Yes |
| 20 | Ikzf4 | 1739 | 0.389 | 0.1677 | Yes |
| 21 | Penk | 1753 | 0.388 | 0.1764 | Yes |
| 22 | Il1r2 | 1820 | 0.383 | 0.1821 | Yes |
| 23 | Serpinc1 | 1888 | 0.380 | 0.1877 | Yes |
| 24 | Anxa4 | 1983 | 0.371 | 0.1917 | Yes |
| 25 | Slc2a3 | 2129 | 0.358 | 0.1928 | Yes |
| 26 | Syt11 | 2287 | 0.343 | 0.1928 | Yes |
| 27 | Ager | 2294 | 0.342 | 0.2007 | Yes |
| 28 | Ptrh2 | 2379 | 0.334 | 0.2044 | Yes |
| 29 | Prkch | 2430 | 0.331 | 0.2097 | Yes |
| 30 | Snx14 | 2487 | 0.327 | 0.2146 | Yes |
| 31 | Plagl1 | 2507 | 0.325 | 0.2214 | Yes |
| 32 | Igf2r | 2560 | 0.321 | 0.2264 | Yes |
| 33 | Tlr7 | 2628 | 0.315 | 0.2305 | Yes |
| 34 | Lclat1 | 2953 | 0.290 | 0.2205 | No |
| 35 | Praf2 | 2994 | 0.287 | 0.2253 | No |
| 36 | Amacr | 3053 | 0.282 | 0.2291 | No |
| 37 | Cd48 | 3220 | 0.273 | 0.2270 | No |
| 38 | Itih5 | 3473 | 0.260 | 0.2201 | No |
| 39 | Cd86 | 3671 | 0.249 | 0.2157 | No |
| 40 | Selp | 3761 | 0.245 | 0.2170 | No |
| 41 | Ncoa3 | 3833 | 0.241 | 0.2191 | No |
| 42 | Tnfrsf21 | 4352 | 0.214 | 0.1971 | No |
| 43 | Slc29a2 | 4418 | 0.210 | 0.1988 | No |
| 44 | Rora | 4443 | 0.208 | 0.2025 | No |
| 45 | Rragd | 4632 | 0.202 | 0.1976 | No |
| 46 | Furin | 4990 | 0.185 | 0.1834 | No |
| 47 | Spred2 | 5290 | 0.171 | 0.1718 | No |
| 48 | Umps | 5296 | 0.171 | 0.1757 | No |
| 49 | Odc1 | 5304 | 0.171 | 0.1794 | No |
| 50 | Cdcp1 | 5321 | 0.170 | 0.1827 | No |
| 51 | Ckap4 | 5409 | 0.167 | 0.1821 | No |
| 52 | Pnp | 5460 | 0.164 | 0.1835 | No |
| 53 | Plec | 5472 | 0.164 | 0.1868 | No |
| 54 | Myo1c | 5542 | 0.161 | 0.1871 | No |
| 55 | Huwe1 | 5581 | 0.159 | 0.1889 | No |
| 56 | Lrrc8c | 5794 | 0.149 | 0.1814 | No |
| 57 | Il18r1 | 5832 | 0.147 | 0.1830 | No |
| 58 | Tgm2 | 6037 | 0.140 | 0.1757 | No |
| 59 | Pth1r | 6061 | 0.139 | 0.1779 | No |
| 60 | Klf6 | 6135 | 0.136 | 0.1773 | No |
| 61 | Hycc2 | 6362 | 0.126 | 0.1685 | No |
| 62 | Gabarapl1 | 7051 | 0.099 | 0.1349 | No |
| 63 | Bcl2 | 7096 | 0.098 | 0.1350 | No |
| 64 | Itgav | 7124 | 0.096 | 0.1359 | No |
| 65 | Ahr | 7140 | 0.096 | 0.1374 | No |
| 66 | Ptger2 | 7226 | 0.092 | 0.1352 | No |
| 67 | Uck2 | 7493 | 0.083 | 0.1233 | No |
| 68 | Pou2f1 | 7725 | 0.073 | 0.1130 | No |
| 69 | Rabgap1l | 7781 | 0.071 | 0.1118 | No |
| 70 | Rhoh | 7904 | 0.067 | 0.1070 | No |
| 71 | Igf1r | 8012 | 0.062 | 0.1029 | No |
| 72 | Nfil3 | 8062 | 0.060 | 0.1018 | No |
| 73 | Tnfrsf18 | 8171 | 0.056 | 0.0975 | No |
| 74 | Csf1 | 8287 | 0.051 | 0.0927 | No |
| 75 | Gata1 | 8393 | 0.047 | 0.0884 | No |
| 76 | Il4ra | 8428 | 0.046 | 0.0877 | No |
| 77 | Il10ra | 8487 | 0.044 | 0.0857 | No |
| 78 | Slc39a8 | 8564 | 0.041 | 0.0827 | No |
| 79 | Bhlhe40 | 8754 | 0.034 | 0.0736 | No |
| 80 | Pdcd2l | 8798 | 0.032 | 0.0722 | No |
| 81 | Hipk2 | 9106 | 0.021 | 0.0566 | No |
| 82 | Alcam | 9159 | 0.019 | 0.0544 | No |
| 83 | Ncs1 | 9245 | 0.016 | 0.0503 | No |
| 84 | Casp3 | 9310 | 0.014 | 0.0473 | No |
| 85 | Ikzf2 | 9403 | 0.011 | 0.0428 | No |
| 86 | Ccnd3 | 9533 | 0.006 | 0.0362 | No |
| 87 | Ptch1 | 9647 | 0.002 | 0.0303 | No |
| 88 | Tnfsf10 | 9649 | 0.002 | 0.0303 | No |
| 89 | Cxcl10 | 10138 | -0.008 | 0.0050 | No |
| 90 | Mapkapk2 | 10380 | -0.017 | -0.0072 | No |
| 91 | Ctla4 | 10406 | -0.018 | -0.0081 | No |
| 92 | Myc | 10474 | -0.020 | -0.0111 | No |
| 93 | P4ha1 | 10495 | -0.021 | -0.0116 | No |
| 94 | Phtf2 | 10502 | -0.021 | -0.0114 | No |
| 95 | F2rl2 | 10692 | -0.028 | -0.0206 | No |
| 96 | Abcb1a | 10784 | -0.031 | -0.0246 | No |
| 97 | Dennd5a | 10874 | -0.034 | -0.0285 | No |
| 98 | Pus1 | 10921 | -0.036 | -0.0300 | No |
| 99 | Gucy1b1 | 11175 | -0.045 | -0.0422 | No |
| 100 | Rhob | 11279 | -0.048 | -0.0464 | No |
| 101 | Glipr2 | 11530 | -0.057 | -0.0581 | No |
| 102 | Cdc6 | 11595 | -0.059 | -0.0600 | No |
| 103 | Cd44 | 11601 | -0.059 | -0.0589 | No |
| 104 | Wls | 11602 | -0.059 | -0.0574 | No |
| 105 | Itga6 | 11690 | -0.063 | -0.0605 | No |
| 106 | Prnp | 11741 | -0.064 | -0.0615 | No |
| 107 | Cish | 12056 | -0.076 | -0.0761 | No |
| 108 | Cdc42se2 | 12106 | -0.078 | -0.0768 | No |
| 109 | Adam19 | 12184 | -0.081 | -0.0789 | No |
| 110 | P2rx4 | 12317 | -0.087 | -0.0837 | No |
| 111 | Spry4 | 12351 | -0.088 | -0.0833 | No |
| 112 | Maff | 12495 | -0.095 | -0.0885 | No |
| 113 | Cdkn1c | 12533 | -0.096 | -0.0881 | No |
| 114 | St3gal4 | 12659 | -0.102 | -0.0922 | No |
| 115 | Gsto1 | 12922 | -0.113 | -0.1032 | No |
| 116 | Swap70 | 12939 | -0.114 | -0.1013 | No |
| 117 | Twsg1 | 13019 | -0.117 | -0.1026 | No |
| 118 | Flt3l | 13040 | -0.118 | -0.1008 | No |
| 119 | Ccnd2 | 13547 | -0.140 | -0.1239 | No |
| 120 | Nfkbiz | 13616 | -0.143 | -0.1240 | No |
| 121 | Irf8 | 13626 | -0.144 | -0.1211 | No |
| 122 | Gpx4 | 13874 | -0.155 | -0.1302 | No |
| 123 | Ifngr1 | 13911 | -0.157 | -0.1283 | No |
| 124 | Itgae | 14078 | -0.165 | -0.1331 | No |
| 125 | Hk2 | 14084 | -0.165 | -0.1293 | No |
| 126 | Ttc39b | 14132 | -0.167 | -0.1278 | No |
| 127 | Eef1akmt1 | 14138 | -0.167 | -0.1240 | No |
| 128 | Bcl2l1 | 14172 | -0.169 | -0.1217 | No |
| 129 | Nop2 | 14181 | -0.169 | -0.1180 | No |
| 130 | Traf1 | 14227 | -0.172 | -0.1163 | No |
| 131 | Fah | 14399 | -0.179 | -0.1209 | No |
| 132 | Car2 | 14624 | -0.189 | -0.1281 | No |
| 133 | Serpinb6a | 14775 | -0.197 | -0.1312 | No |
| 134 | Gpr65 | 14821 | -0.199 | -0.1288 | No |
| 135 | Snx9 | 15098 | -0.214 | -0.1380 | No |
| 136 | Dcps | 15172 | -0.218 | -0.1366 | No |
| 137 | Ndrg1 | 15239 | -0.222 | -0.1347 | No |
| 138 | Il3ra | 15362 | -0.229 | -0.1356 | No |
| 139 | Bmp2 | 15795 | -0.248 | -0.1523 | No |
| 140 | Dhrs3 | 15803 | -0.248 | -0.1467 | No |
| 141 | Nrp1 | 15970 | -0.259 | -0.1491 | No |
| 142 | Bmpr2 | 16041 | -0.264 | -0.1464 | No |
| 143 | Myo1e | 16134 | -0.269 | -0.1448 | No |
| 144 | Socs1 | 16201 | -0.273 | -0.1417 | No |
| 145 | Ltb | 16202 | -0.273 | -0.1351 | No |
| 146 | Map3k8 | 16223 | -0.274 | -0.1295 | No |
| 147 | Drc1 | 16240 | -0.276 | -0.1237 | No |
| 148 | Cyfip1 | 16329 | -0.281 | -0.1216 | No |
| 149 | Cst7 | 16400 | -0.287 | -0.1183 | No |
| 150 | Rnh1 | 16456 | -0.290 | -0.1142 | No |
| 151 | Il10 | 16748 | -0.307 | -0.1221 | No |
| 152 | Plpp1 | 16794 | -0.311 | -0.1169 | No |
| 153 | Xbp1 | 16850 | -0.316 | -0.1122 | No |
| 154 | Etfbkmt | 16879 | -0.318 | -0.1060 | No |
| 155 | Arl4a | 16973 | -0.324 | -0.1031 | No |
| 156 | Galm | 17156 | -0.338 | -0.1045 | No |
| 157 | Icos | 17343 | -0.353 | -0.1057 | No |
| 158 | Socs2 | 17405 | -0.358 | -0.1003 | No |
| 159 | Fgl2 | 17452 | -0.363 | -0.0940 | No |
| 160 | Cd79b | 17465 | -0.364 | -0.0859 | No |
| 161 | Slc1a5 | 17468 | -0.364 | -0.0772 | No |
| 162 | Plscr1 | 17524 | -0.370 | -0.0712 | No |
| 163 | Capg | 17553 | -0.372 | -0.0637 | No |
| 164 | Enpp1 | 17649 | -0.382 | -0.0595 | No |
| 165 | Smpdl3a | 17660 | -0.382 | -0.0508 | No |
| 166 | Cd81 | 17694 | -0.386 | -0.0433 | No |
| 167 | Ctsz | 17972 | -0.419 | -0.0477 | No |
| 168 | S100a1 | 18183 | -0.446 | -0.0480 | No |
| 169 | Mxd1 | 18294 | -0.457 | -0.0427 | No |
| 170 | Syngr2 | 18448 | -0.481 | -0.0391 | No |
| 171 | Eno3 | 18489 | -0.490 | -0.0295 | No |
| 172 | Batf | 18771 | -0.545 | -0.0310 | No |
| 173 | Nt5e | 18786 | -0.550 | -0.0186 | No |
| 174 | Plin2 | 18984 | -0.613 | -0.0141 | No |
| 175 | Gbp3 | 18996 | -0.617 | 0.0001 | No |
| 176 | Ifitm3 | 19101 | -0.669 | 0.0108 | No |