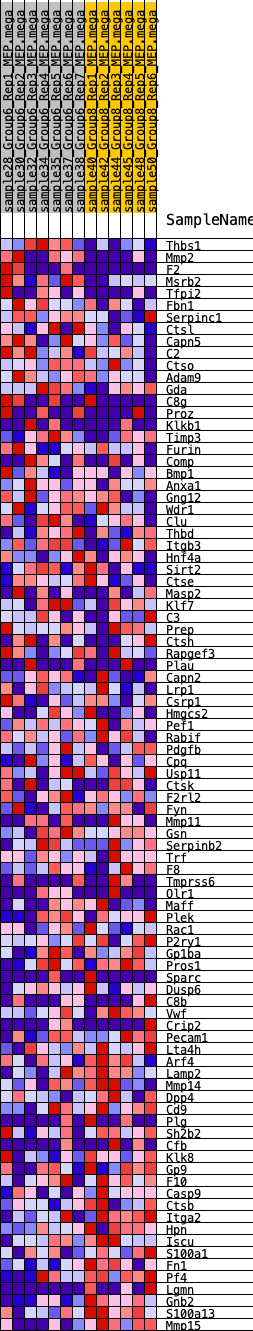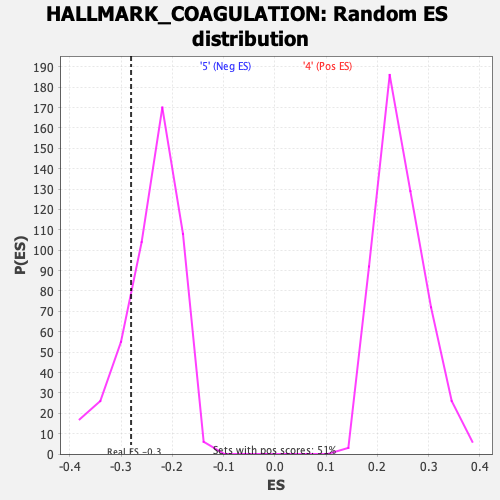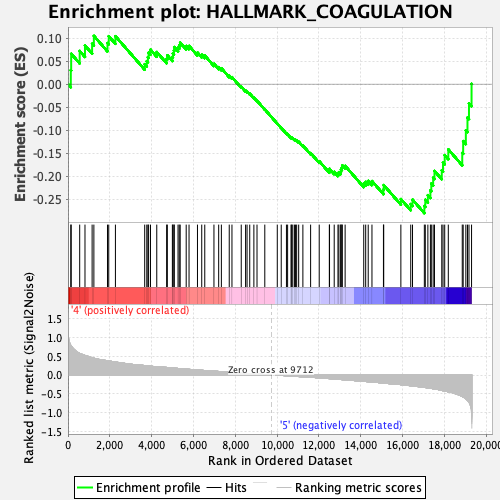
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group6_versus_Group8.MEP.mega_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.mega_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.28055874 |
| Normalized Enrichment Score (NES) | -1.1718328 |
| Nominal p-value | 0.20164609 |
| FDR q-value | 0.3991613 |
| FWER p-Value | 0.955 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thbs1 | 134 | 0.783 | 0.0305 | No |
| 2 | Mmp2 | 152 | 0.763 | 0.0661 | No |
| 3 | F2 | 559 | 0.568 | 0.0722 | No |
| 4 | Msrb2 | 812 | 0.519 | 0.0839 | No |
| 5 | Tfpi2 | 1153 | 0.458 | 0.0881 | No |
| 6 | Fbn1 | 1237 | 0.444 | 0.1050 | No |
| 7 | Serpinc1 | 1888 | 0.380 | 0.0894 | No |
| 8 | Ctsl | 1948 | 0.376 | 0.1043 | No |
| 9 | Capn5 | 2267 | 0.346 | 0.1042 | No |
| 10 | C2 | 3667 | 0.249 | 0.0433 | No |
| 11 | Ctso | 3767 | 0.245 | 0.0499 | No |
| 12 | Adam9 | 3821 | 0.242 | 0.0587 | No |
| 13 | Gda | 3854 | 0.240 | 0.0685 | No |
| 14 | C8g | 3944 | 0.233 | 0.0751 | No |
| 15 | Proz | 4244 | 0.217 | 0.0699 | No |
| 16 | Klkb1 | 4716 | 0.200 | 0.0549 | No |
| 17 | Timp3 | 4749 | 0.197 | 0.0627 | No |
| 18 | Furin | 4990 | 0.185 | 0.0591 | No |
| 19 | Comp | 5015 | 0.184 | 0.0666 | No |
| 20 | Bmp1 | 5065 | 0.181 | 0.0727 | No |
| 21 | Anxa1 | 5079 | 0.181 | 0.0807 | No |
| 22 | Gng12 | 5261 | 0.173 | 0.0795 | No |
| 23 | Wdr1 | 5318 | 0.170 | 0.0848 | No |
| 24 | Clu | 5364 | 0.169 | 0.0905 | No |
| 25 | Thbd | 5651 | 0.155 | 0.0830 | No |
| 26 | Itgb3 | 5787 | 0.149 | 0.0832 | No |
| 27 | Hnf4a | 6190 | 0.133 | 0.0686 | No |
| 28 | Sirt2 | 6394 | 0.125 | 0.0640 | No |
| 29 | Ctse | 6539 | 0.118 | 0.0621 | No |
| 30 | Masp2 | 6978 | 0.101 | 0.0442 | No |
| 31 | Klf7 | 7204 | 0.093 | 0.0369 | No |
| 32 | C3 | 7333 | 0.089 | 0.0345 | No |
| 33 | Prep | 7709 | 0.074 | 0.0185 | No |
| 34 | Ctsh | 7839 | 0.069 | 0.0151 | No |
| 35 | Rapgef3 | 8283 | 0.052 | -0.0054 | No |
| 36 | Plau | 8491 | 0.043 | -0.0141 | No |
| 37 | Capn2 | 8561 | 0.041 | -0.0158 | No |
| 38 | Lrp1 | 8689 | 0.036 | -0.0207 | No |
| 39 | Csrp1 | 8887 | 0.029 | -0.0295 | No |
| 40 | Hmgcs2 | 9037 | 0.024 | -0.0361 | No |
| 41 | Pef1 | 9409 | 0.011 | -0.0549 | No |
| 42 | Rabif | 10006 | -0.003 | -0.0858 | No |
| 43 | Pdgfb | 10195 | -0.010 | -0.0951 | No |
| 44 | Cpq | 10444 | -0.019 | -0.1071 | No |
| 45 | Usp11 | 10500 | -0.021 | -0.1090 | No |
| 46 | Ctsk | 10663 | -0.027 | -0.1161 | No |
| 47 | F2rl2 | 10692 | -0.028 | -0.1163 | No |
| 48 | Fyn | 10746 | -0.030 | -0.1176 | No |
| 49 | Mmp11 | 10826 | -0.032 | -0.1202 | No |
| 50 | Gsn | 10880 | -0.034 | -0.1213 | No |
| 51 | Serpinb2 | 10932 | -0.036 | -0.1222 | No |
| 52 | Trf | 11029 | -0.040 | -0.1253 | No |
| 53 | F8 | 11230 | -0.046 | -0.1335 | No |
| 54 | Tmprss6 | 11600 | -0.059 | -0.1499 | No |
| 55 | Olr1 | 12011 | -0.074 | -0.1677 | No |
| 56 | Maff | 12495 | -0.095 | -0.1883 | No |
| 57 | Plek | 12504 | -0.095 | -0.1842 | No |
| 58 | Rac1 | 12728 | -0.105 | -0.1908 | No |
| 59 | P2ry1 | 12908 | -0.112 | -0.1947 | No |
| 60 | Gp1ba | 12952 | -0.114 | -0.1915 | No |
| 61 | Pros1 | 13038 | -0.118 | -0.1903 | No |
| 62 | Sparc | 13052 | -0.119 | -0.1853 | No |
| 63 | Dusp6 | 13094 | -0.121 | -0.1816 | No |
| 64 | C8b | 13113 | -0.121 | -0.1768 | No |
| 65 | Vwf | 13250 | -0.126 | -0.1778 | No |
| 66 | Crip2 | 14140 | -0.167 | -0.2160 | No |
| 67 | Pecam1 | 14233 | -0.172 | -0.2126 | No |
| 68 | Lta4h | 14354 | -0.177 | -0.2104 | No |
| 69 | Arf4 | 14533 | -0.185 | -0.2108 | No |
| 70 | Lamp2 | 15082 | -0.213 | -0.2291 | No |
| 71 | Mmp14 | 15095 | -0.214 | -0.2195 | No |
| 72 | Dpp4 | 15915 | -0.255 | -0.2499 | No |
| 73 | Cd9 | 16387 | -0.286 | -0.2608 | No |
| 74 | Plg | 16472 | -0.291 | -0.2512 | No |
| 75 | Sh2b2 | 17037 | -0.327 | -0.2649 | Yes |
| 76 | Cfb | 17083 | -0.331 | -0.2514 | Yes |
| 77 | Klk8 | 17206 | -0.341 | -0.2415 | Yes |
| 78 | Gp9 | 17327 | -0.352 | -0.2309 | Yes |
| 79 | F10 | 17370 | -0.356 | -0.2161 | Yes |
| 80 | Casp9 | 17459 | -0.364 | -0.2032 | Yes |
| 81 | Ctsb | 17522 | -0.369 | -0.1888 | Yes |
| 82 | Itga2 | 17871 | -0.408 | -0.1874 | Yes |
| 83 | Hpn | 17930 | -0.414 | -0.1706 | Yes |
| 84 | Iscu | 18011 | -0.426 | -0.1544 | Yes |
| 85 | S100a1 | 18183 | -0.446 | -0.1420 | Yes |
| 86 | Fn1 | 18850 | -0.567 | -0.1495 | Yes |
| 87 | Pf4 | 18894 | -0.581 | -0.1239 | Yes |
| 88 | Lgmn | 19022 | -0.628 | -0.1005 | Yes |
| 89 | Gnb2 | 19100 | -0.668 | -0.0726 | Yes |
| 90 | S100a13 | 19172 | -0.720 | -0.0418 | Yes |
| 91 | Mmp15 | 19295 | -1.020 | 0.0006 | Yes |