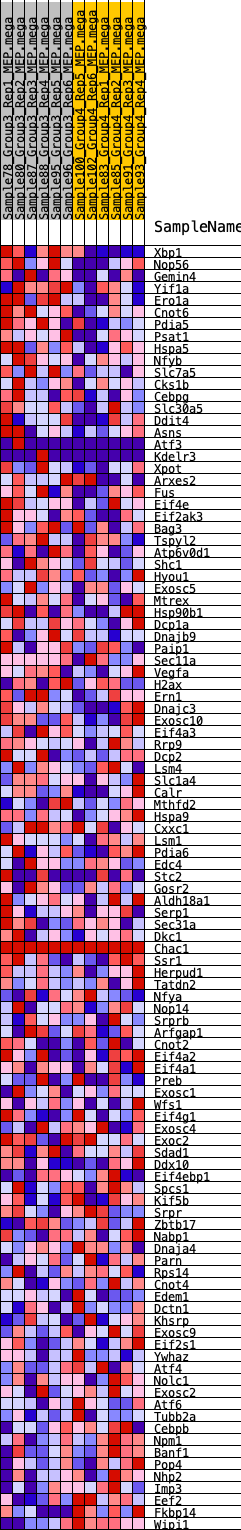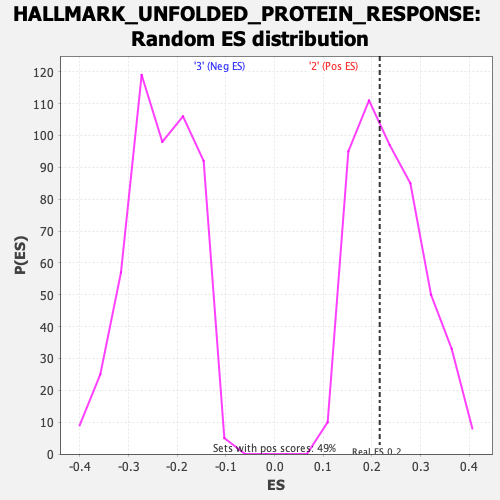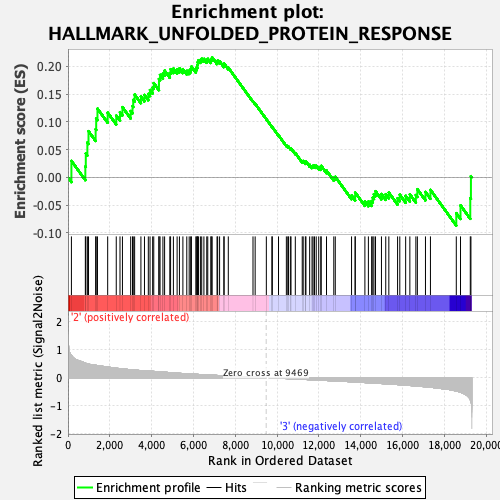
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.2160069 |
| Normalized Enrichment Score (NES) | 0.91884273 |
| Nominal p-value | 0.5460123 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xbp1 | 163 | 0.807 | 0.0297 | Yes |
| 2 | Nop56 | 829 | 0.521 | 0.0197 | Yes |
| 3 | Gemin4 | 851 | 0.516 | 0.0430 | Yes |
| 4 | Yif1a | 926 | 0.502 | 0.0629 | Yes |
| 5 | Ero1a | 981 | 0.489 | 0.0832 | Yes |
| 6 | Cnot6 | 1321 | 0.446 | 0.0866 | Yes |
| 7 | Pdia5 | 1348 | 0.442 | 0.1062 | Yes |
| 8 | Psat1 | 1409 | 0.437 | 0.1238 | Yes |
| 9 | Hspa5 | 1897 | 0.383 | 0.1165 | Yes |
| 10 | Nfyb | 2307 | 0.340 | 0.1113 | Yes |
| 11 | Slc7a5 | 2486 | 0.326 | 0.1175 | Yes |
| 12 | Cks1b | 2603 | 0.318 | 0.1264 | Yes |
| 13 | Cebpg | 2997 | 0.287 | 0.1195 | Yes |
| 14 | Slc30a5 | 3084 | 0.280 | 0.1283 | Yes |
| 15 | Ddit4 | 3119 | 0.277 | 0.1396 | Yes |
| 16 | Asns | 3186 | 0.273 | 0.1491 | Yes |
| 17 | Atf3 | 3486 | 0.255 | 0.1456 | Yes |
| 18 | Kdelr3 | 3656 | 0.246 | 0.1484 | Yes |
| 19 | Xpot | 3840 | 0.242 | 0.1504 | Yes |
| 20 | Arxes2 | 3922 | 0.237 | 0.1573 | Yes |
| 21 | Fus | 4047 | 0.230 | 0.1618 | Yes |
| 22 | Eif4e | 4100 | 0.227 | 0.1698 | Yes |
| 23 | Eif2ak3 | 4348 | 0.215 | 0.1671 | Yes |
| 24 | Bag3 | 4349 | 0.215 | 0.1772 | Yes |
| 25 | Tspyl2 | 4401 | 0.211 | 0.1845 | Yes |
| 26 | Atp6v0d1 | 4538 | 0.203 | 0.1871 | Yes |
| 27 | Shc1 | 4619 | 0.198 | 0.1922 | Yes |
| 28 | Hyou1 | 4868 | 0.183 | 0.1880 | Yes |
| 29 | Exosc5 | 4905 | 0.182 | 0.1947 | Yes |
| 30 | Mtrex | 5039 | 0.174 | 0.1960 | Yes |
| 31 | Hsp90b1 | 5217 | 0.164 | 0.1945 | Yes |
| 32 | Dcp1a | 5328 | 0.158 | 0.1963 | Yes |
| 33 | Dnajb9 | 5493 | 0.150 | 0.1949 | Yes |
| 34 | Paip1 | 5679 | 0.142 | 0.1919 | Yes |
| 35 | Sec11a | 5794 | 0.137 | 0.1924 | Yes |
| 36 | Vegfa | 5866 | 0.133 | 0.1951 | Yes |
| 37 | H2ax | 5900 | 0.132 | 0.1996 | Yes |
| 38 | Ern1 | 6112 | 0.124 | 0.1944 | Yes |
| 39 | Dnajc3 | 6151 | 0.122 | 0.1982 | Yes |
| 40 | Exosc10 | 6186 | 0.121 | 0.2022 | Yes |
| 41 | Eif4a3 | 6198 | 0.120 | 0.2073 | Yes |
| 42 | Rrp9 | 6237 | 0.118 | 0.2109 | Yes |
| 43 | Dcp2 | 6330 | 0.115 | 0.2116 | Yes |
| 44 | Lsm4 | 6393 | 0.112 | 0.2137 | Yes |
| 45 | Slc1a4 | 6498 | 0.108 | 0.2134 | Yes |
| 46 | Calr | 6634 | 0.103 | 0.2112 | Yes |
| 47 | Mthfd2 | 6682 | 0.101 | 0.2135 | Yes |
| 48 | Hspa9 | 6828 | 0.096 | 0.2105 | Yes |
| 49 | Cxxc1 | 6853 | 0.094 | 0.2137 | Yes |
| 50 | Lsm1 | 6893 | 0.092 | 0.2160 | Yes |
| 51 | Pdia6 | 7126 | 0.083 | 0.2078 | No |
| 52 | Edc4 | 7155 | 0.082 | 0.2102 | No |
| 53 | Stc2 | 7248 | 0.079 | 0.2092 | No |
| 54 | Gosr2 | 7455 | 0.073 | 0.2019 | No |
| 55 | Aldh18a1 | 7461 | 0.072 | 0.2050 | No |
| 56 | Serp1 | 7664 | 0.063 | 0.1975 | No |
| 57 | Sec31a | 8845 | 0.020 | 0.1370 | No |
| 58 | Dkc1 | 8948 | 0.018 | 0.1326 | No |
| 59 | Chac1 | 9483 | 0.000 | 0.1048 | No |
| 60 | Ssr1 | 9748 | -0.004 | 0.0912 | No |
| 61 | Herpud1 | 9769 | -0.005 | 0.0904 | No |
| 62 | Tatdn2 | 10065 | -0.015 | 0.0757 | No |
| 63 | Nfya | 10440 | -0.028 | 0.0576 | No |
| 64 | Nop14 | 10509 | -0.030 | 0.0555 | No |
| 65 | Srprb | 10541 | -0.031 | 0.0553 | No |
| 66 | Arfgap1 | 10631 | -0.034 | 0.0523 | No |
| 67 | Cnot2 | 10662 | -0.035 | 0.0524 | No |
| 68 | Eif4a2 | 10868 | -0.043 | 0.0438 | No |
| 69 | Eif4a1 | 11199 | -0.055 | 0.0292 | No |
| 70 | Preb | 11245 | -0.057 | 0.0295 | No |
| 71 | Exosc1 | 11348 | -0.060 | 0.0271 | No |
| 72 | Wfs1 | 11376 | -0.061 | 0.0286 | No |
| 73 | Eif4g1 | 11554 | -0.069 | 0.0226 | No |
| 74 | Exosc4 | 11668 | -0.073 | 0.0202 | No |
| 75 | Exoc2 | 11717 | -0.074 | 0.0212 | No |
| 76 | Sdad1 | 11785 | -0.077 | 0.0213 | No |
| 77 | Ddx10 | 11868 | -0.080 | 0.0208 | No |
| 78 | Eif4ebp1 | 11987 | -0.083 | 0.0186 | No |
| 79 | Spcs1 | 12087 | -0.087 | 0.0176 | No |
| 80 | Kif5b | 12105 | -0.087 | 0.0208 | No |
| 81 | Srpr | 12360 | -0.098 | 0.0122 | No |
| 82 | Zbtb17 | 12708 | -0.111 | -0.0006 | No |
| 83 | Nabp1 | 12779 | -0.114 | 0.0012 | No |
| 84 | Dnaja4 | 13557 | -0.144 | -0.0325 | No |
| 85 | Parn | 13730 | -0.150 | -0.0343 | No |
| 86 | Rps14 | 13731 | -0.151 | -0.0272 | No |
| 87 | Cnot4 | 14195 | -0.171 | -0.0432 | No |
| 88 | Edem1 | 14362 | -0.179 | -0.0434 | No |
| 89 | Dctn1 | 14517 | -0.186 | -0.0426 | No |
| 90 | Khsrp | 14572 | -0.189 | -0.0365 | No |
| 91 | Exosc9 | 14627 | -0.192 | -0.0302 | No |
| 92 | Eif2s1 | 14703 | -0.195 | -0.0249 | No |
| 93 | Ywhaz | 14987 | -0.206 | -0.0299 | No |
| 94 | Atf4 | 15192 | -0.216 | -0.0303 | No |
| 95 | Nolc1 | 15340 | -0.223 | -0.0274 | No |
| 96 | Exosc2 | 15758 | -0.243 | -0.0376 | No |
| 97 | Atf6 | 15864 | -0.251 | -0.0312 | No |
| 98 | Tubb2a | 16144 | -0.265 | -0.0332 | No |
| 99 | Cebpb | 16345 | -0.276 | -0.0306 | No |
| 100 | Npm1 | 16632 | -0.292 | -0.0316 | No |
| 101 | Banf1 | 16704 | -0.297 | -0.0213 | No |
| 102 | Pop4 | 17091 | -0.322 | -0.0262 | No |
| 103 | Nhp2 | 17329 | -0.339 | -0.0225 | No |
| 104 | Imp3 | 18565 | -0.473 | -0.0644 | No |
| 105 | Eef2 | 18767 | -0.519 | -0.0504 | No |
| 106 | Fkbp14 | 19234 | -0.790 | -0.0373 | No |
| 107 | Wipi1 | 19265 | -0.867 | 0.0022 | No |